Figures & data
Figure 1. Irradiated E1A + E1B cells arrest cell cycle progression in G2/M phase and suppress proliferation while continue to replicate DNA. (A) Cell cycle distribution analyzed by flow cytometry of propidium iodide-stained cells. Percentage of cells in S phase and percent of polyploid cells are shown. (B) Analysis of DNA-replication in cells according to BrdU incorporation. Non-irradiated and IR-treated cells were pulse-labeled with BrdU for 1 h, followed by immunofluorescent staining. (C) Growth curves of irradiated and untreated E1A + E1B cells. Cells were seeded in initial density of 3 × 104 cells per 30-mm dish and counted daily. Mean data with standard deviation are shown.
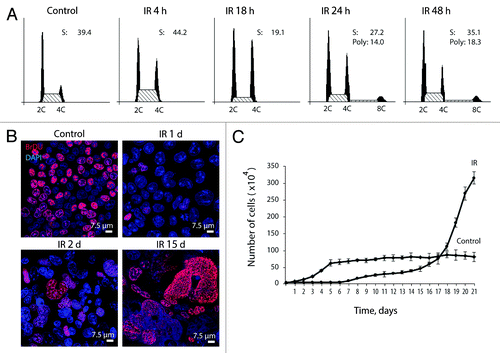
Figure 2. Exposure of E1A + E1B cells to IR results in the formation of giant polyploid cells, which are characterized by high level of E1A protein expression. (A) Microphotographs of cells stained with hematoxylin and eosin. Images were acquired in transmitted light, magnification 10 × 40. (B) Frequency distribution of cells according to DNA content was calculated by DNA cytometry of Feulgen-stained samples. Analysis of E1A expression in non-irradiated and IR-exposed cells by western blot (C) and immunofluorescent staining (D).
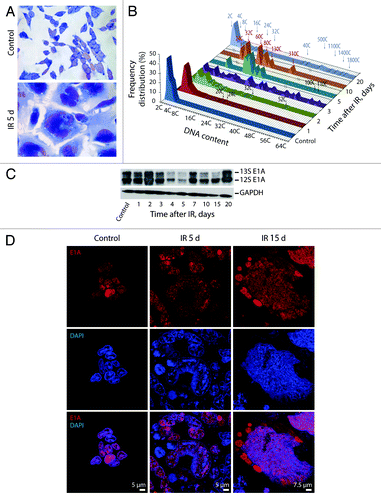
Figure 3. Kinetics of γH2AX and 53BP1 foci formation and resolution in E1A + E1B cells. (A) Colocalization and persistence of γH2AX and 53BP1 foci in E1A + E1B cells after exposure to IR. Cells were irradiated or left untreated and stained with antibodies against γH2AX and 53BP1. Confocal images are shown. (B) Number of γH2AX foci per cell in E1A + E1B cells and REFs. (C) The percentage of cells with γH2AX foci. (D) Number of 53BP1 foci per cell in E1A + E1B cells and REFs. (E) The percentage of cells with 53BP1 foci. Note for (B) and (D): only cells with foci were included in the analysis. Note for (C) and (E): untreated cells contain 0–3 DDR foci per cell; therefore, cells with more than 3 foci were counted. (B–E) Mean data with the standard deviation are shown.
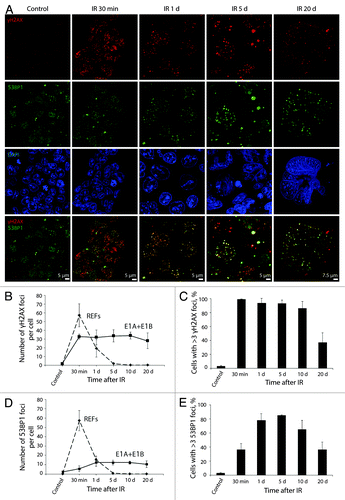
Figure 4. pATMSer1981 is a component of early and persistent DDR foci. E1A + E1B cells were irradiated or left untreated and stained with the antibodies against pATMSer1981 and 53BP1. Colocalization of pATMSer1981 and 53BP1 in giant cell is indicated with arrows. Confocal images are shown.
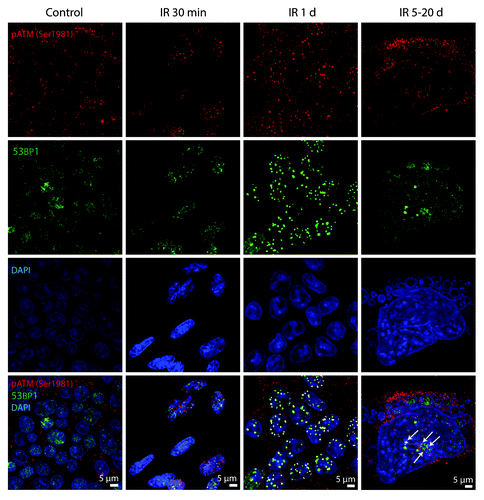
Figure 5. pATRSer428 does not colocolize with DDR foci in E1A + E1B cells. Irradiated and untreated cells were stained with the antibodies against pATRSer428 and γH2AX. Confocal images are shown.
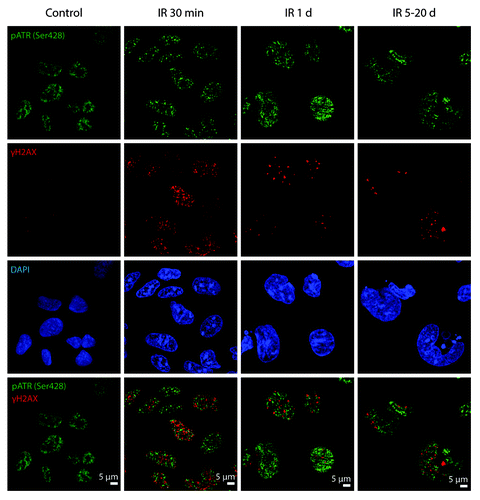
Figure 6. Analysis of DNA breaks persistence in E1A + E1B cells. (A) Untreated and irradiated cells were subjected to single-cell gel electrophoresis at the indicated time intervals after exposure to IR. Magnification 10 × 20. (B) Quantification of percentage of cells with DNA breaks in untreated and irradiated cells. Measurement of comet tail length (C), and comet tail moment (D), performed with CaspLab software. (E) Quantification of percentage of viable cells based on acridin orange and ethidium bromide staining. Mean data with standard deviation are shown for (B–E).
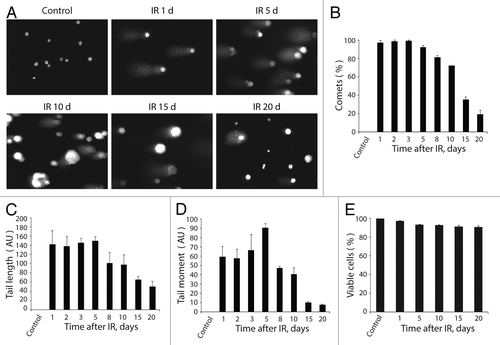
Figure 7. Irradiated E1A + E1B cells show delayed accumulation and persistence of Rad51 within the DDR foci. (A) Cells were left untreated or irradiated followed by staining with antibodies against Rad51 and γH2AX. Confocal images are shown. (B) Fluorescence intensity of Rad51 in untreated and irradiated cells was calculated as ratio of raw density to the cell surface measured with ImageJ software. Only cells expressing Rad51 were included in the analysis. (C) The percentage of cells containing Rad51 foci. (B and C) Mean data with standard deviation are shown. (D) Colocalization of Rad51 and γH2AX in the micronuclei indicate elimination of damaged DNA. Confocal images are shown.
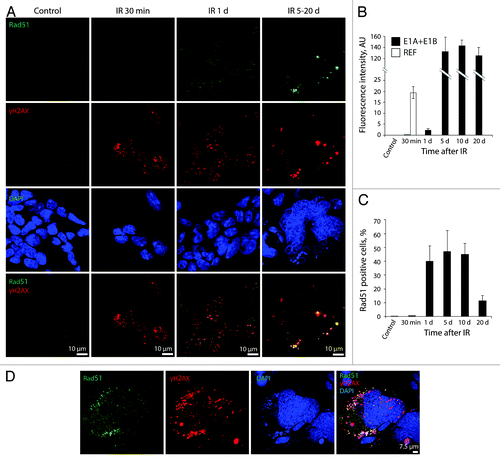
Figure 8. pDNA-PKcsSer2056 colocolizes with DDR foci within the minutes after irradiation and remains persistent. (A) Cells were irradiated or left untreated and stained with antibodies against pDNA-PKcsSer2056 and γH2AX. Confocal images are shown. (B) Fluorescence intensity of pDNA-PKcsSer2056 in untreated and irradiated cells was calculated as ratio of raw density to the cell surface measured with ImageJ software. Only cells that express pDNA-PKcsSer2056 were included in the analysis. (C) The percentage of cells containing pDNA-PKcsSer2056 foci. (B and C) Mean data with standard deviation are shown.
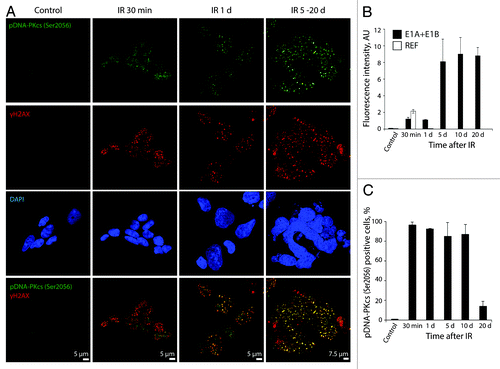
Figure 9. Analysis of colocalization of DDR foci with the sites of DNA replication. Non-irradiated and IR-exposed cells were subjected to EdU incorporation assay by “click-it” method and stained with antibodies against γH2AX. Confocal images are shown.
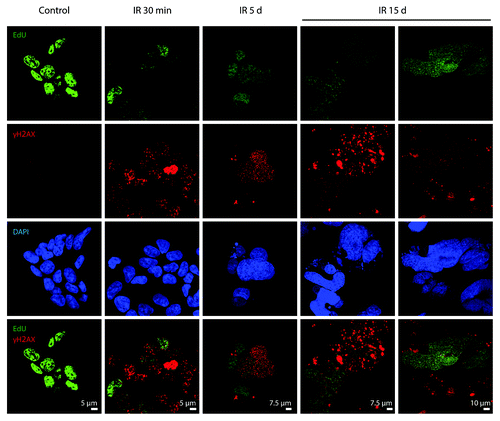
Figure 10. E1A + E1B cells overpass senescence induced by IR. (A) SA-β-Gal staining of untreated and irradiated cells was performed. Images were acquired in transmitted light, magnification 10 × 40. Giant cells remain SA-β-Gal-positive (a), whereas cells of near-normal size are SA-β-Gal-negative (b). (B) Quantification of the percentage of senescent cells stained for SA-β-Gal detection. Mean values with standard deviation are shown.
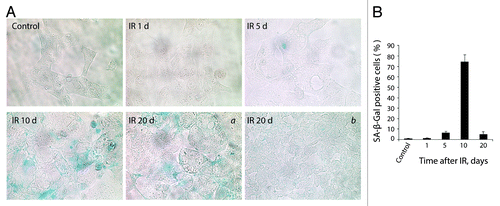
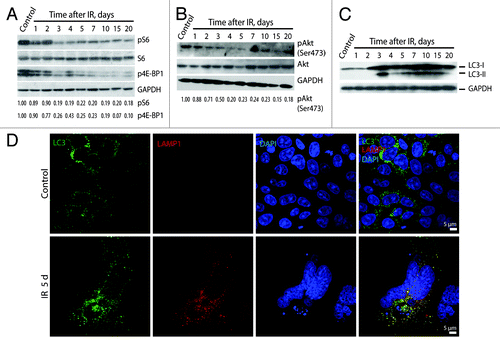
Figure 11. Irradiated E1A + E1B cells show suppression of mTORC1 and mTORC2 and activation of autophagy. Western blot analysis of phosphorylation of S6 ribosomal protein and 4E-BP1 (A) and phosphorylation of Akt on Ser473 (B). The indicated numbers show the results of western blot densitometry. (C) Western blot analysis of LC3-I conversion to LC3-II. (D) Analysis of LC3 and LAMP1 colocalization in non-irradiated and IR-treated cells. Confocal images are shown.