Figures & data
Figure 1. EGFP-LactC2 (PS) and mCherry-Rab GTPases in live HeLa cells show distinct patterns of overlap. HeLa cells expressing EGFP-LactC2 and mCherry-Rab proteins were imaged with live cell deconvolution microscopy for at least 1 min every 2 s. Rab5a and Rab7a showed limited overlap with LactC2 in the periphery of cells while Rab8a and Rab11a display consistent overlap with mCherry-LactC2 throughout the cell. Rab10 overlap with LactC2 was present but not consistent over multiple experiments. Data represent at least 3 independent experiments. Bars, 10 μm
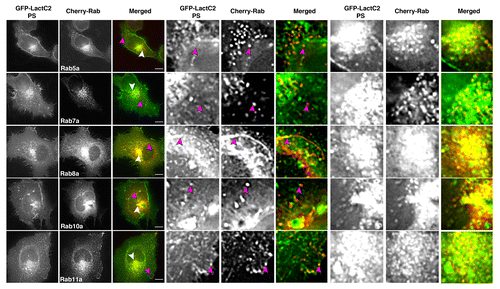
Figure 2. EGFP-Rab11-FIP1 proteins consistently overlap with mCherry-LactC2 (PS) in live HeLa cells. EGFP-Rab11-FIP1 proteins and mCherry-LactC2 overlapped in peripheral and pericentriolar compartments during imaging of live HeLa cells. FIP1B and FIP1C induced a partial accumulation of LactC2 in the pericentriolar compartments. Cells were imaged for at least one minute every 2 s. Data represent at least 3 independent experiments. Bars, 10 μm.
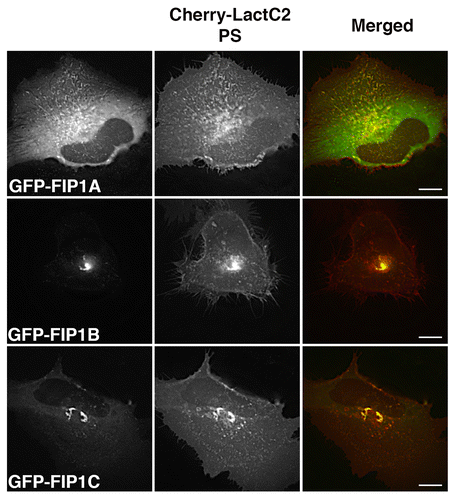
Figure 3. The Rab11-FIP1 proteins are within 100–200nm of LactC2 by SIM. (A) HeLa cells transfected with EGFP-Rab11-FIP1 proteins and mCherry-LactC2 were imaged on coverslips using structured illumination microscopy. Each Rab11-FIP1 protein displayed overlap with LactC2. Images were collected over a 1 μm stack of individual HeLa cells. Bars, 10 μm. (B) Pearson’s correlation coefficients were analyzed for each condition. Rab11-FIP1A (0.472 ± 0.029, n = 14 cells), Rab11-FIP1B (0.491 ± 0.044, n = 9 cells), and Rab11-FIP1C (0.462 ± 0.045, n = 8 cells) had statistically similar overlap with LactC2 (P > 0.05) Results were analyzed using an unpaired, two-tailed, Student’s t test and presented as Mean ± SEM.
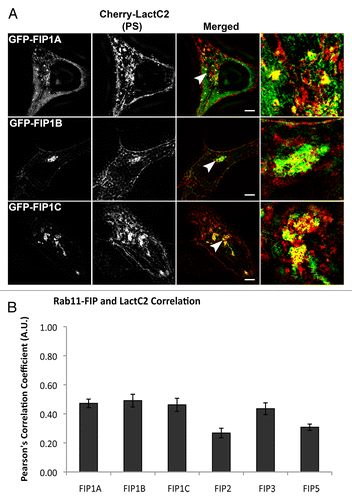
Figure 4. Alternative EGFP-Rab11-FIPs exhibit selective overlap with mCherry-LactC2. HeLa cells coexpressing EGFP-Rab11-FIPs and mCherry-LactC2 were imaged using live cell deconvolution microscopy. Images were collected every 2 s for at least 1 min. EGFP-Rab11-FIP2 and EGFP-Rab11-FIP5 were separate from mCherry LactC2 particularly in the periphery of HeLa cells. EGFP-Rab11-FIP3 was overlapped with mCherry-LactC2 in the pericentriolar region of the cell on distinct tubular membranes. Data represent at least 3 independent experiments. Bars,10 μm.
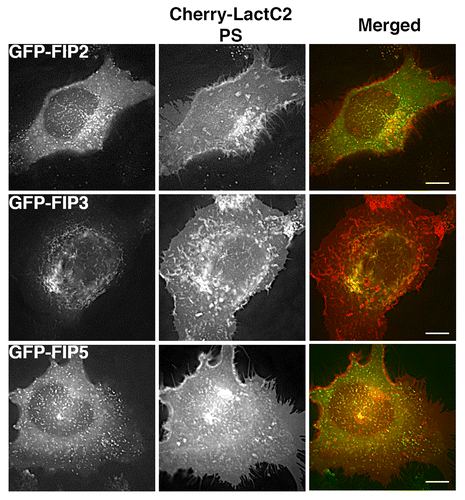
Figure 5: mCherry-LactC2 with alternative EGFP-Rab11-FIPs is limited to EGFP-FIP3 within the pericentriolar region by SIM. A. EGFP-Rab11-FIPs encoded by genes other than Rab11-FIP1 do not overlap as extensively with LactC2 in HeLa cells. HeLa cells expressing EGFP-Rab11-FIPs and mCherry-LactC2 were imaged on coverslips by structured illumination microscopy. EGFP-Rab11-FIP3 was observed along mCherry-LactC2 positive compartments within the perincentriolar region while EGFP-Rab11-FIP2 and EGFP-Rab11-FIP5 did not overlap with mCherry-LactC2. Bars,10 μm. B. Pearson’s correlation coefficients were analyzed for each condition. Rab11-FIP2 (0.268 ± 0.033, n = 8 cells) and Rab11-FIP5 (0.308 ± 0.022, n = 10 cells) had significantly lower correlation coefficients (P < 0.05) than other Rab11-FIPs. Rab11-FIP3 (0.435 ± 0.041, n = 17 cells; P > 0.05) was not significantly different than Rab11-FIP1 proteins. Results were analyzed using an unpaired, two-tailed, Student’s t test and presented as Mean ± SEM.
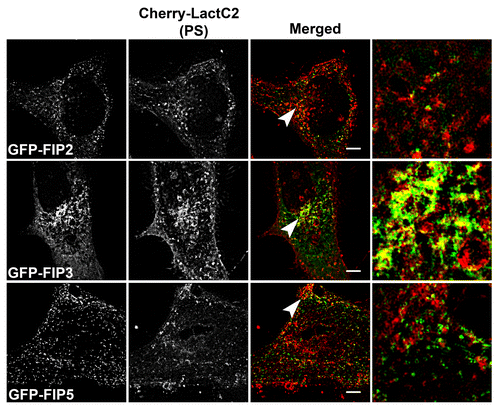
Figure 6. mCherry-LactC2 is enriched along tubular compartments of the Rab11-FIP network. Cerulean-Rab11-FIP3, Venus-Rab11-FIP1A, and mCherry-Rab11-LactC2 expressed in HeLa cells overlap along endosomal tubules in the pericentriolar region. Images were collected every 3 s for at least 1 min. Bars, 10μm.
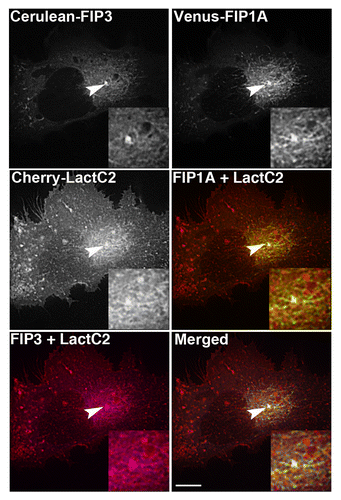
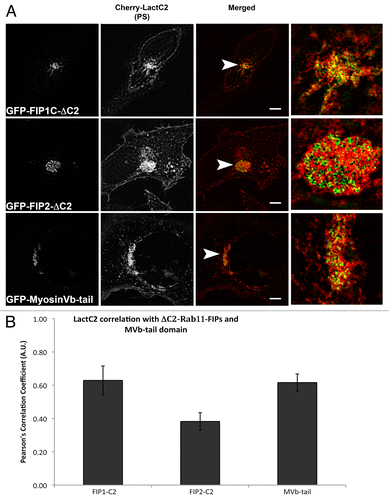
Figure 7. EGFP-Rab11-FIPs lacking the N-terminal C2 domain induce accumulation of LactC2 in the pericentriolar region. EGFP-Rab11-FIP1CΔC2 or EGFP-Rab11-FIP2ΔC2 cause an accumulation of mCherry-LactC2 in the pericentriolar region of live HeLa cells. Expression of MyosinVb-tail induced a similar accumulation of mCherry-LactC2. Images were collected every 2 s for at least 1 min. Data are representative of 2 independent experiments. Bar, 10μm.
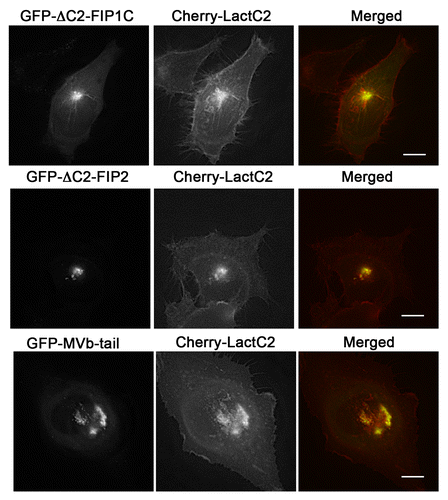
Figure 8: The mCherry-LactC2 and Rab11-FIP-ΔC2 proteins exhibit selective overlap in the pericentriolar region. (A) EGFP-Rab11-FIPΔC2 constructs coexpressed with mCherry-LactC2 on coverslips and imaged using structured illumination microscopy demonstrated distinct associations between LactC2 and Rab11-FIP1C or Myosin Vb, but visible separation between LactC2 and Rab11-FIP2 . Bars, 10 μm. (B) ΔC2-Rab11-FIP2 (0.383 ± 0.052, n = 9 cells) and LactC2 (P < 0.05) had significantly lower correlation than ΔC2-Rab11-FIP1C (0.629 ± 0.086, n = 3 cells) or Myosin Vb tail (0.616 ± 0.051, n = 9 cells; P > 0.05). Results were analyzed using an unpaired, two-tailed, Student’s t test and presented as Mean ± SEM.