Figures & data
Figure 1. Epigenetic regulation of the immune response in transplantation. (A) DNA methylation and histone modifications allow changes in chromatin structure that influence gene transcription. DNA methylation (black circles) and repressive histone marks (H3K27me3, H3K9me3, etc.) are associated with closed chromatin and gene repression while DNA demethylation (white circles) and active histone modifications (AcH3, AcH4, H3K4me3, H3K36me3, etc.) are correlated with an open chromatin structure, facilitating transcription factor binding and transcription. Enzymes such as histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs), histone demethylases (HDMTs) and DNA methyltransferases (DNMTs), establish and maintain the balance between these modifications. (B) Changes in the chromatin structure of key genes in the immune responses that take place in organ transplantation can contribute to alloimmunity or tolerance. Epigenetic modifications (DNA methylation and histone modifications) regulate gene expression in different cell types in several ways: (1) the expression of pro-inflammatory (IL12, IL-1β, IL-6, TNF-α), regulatory (IL-10) cytokines, and co-stimulatory molecules (CD80, CD86, CD40) is modified in antigen-presenting cells (APCs); (2) DNA demethylation and histone deacetylation increase the stable expression of the FoxP3 transcription factor, strengthening the suppressive function of Treg cells; (3) in CD4+ T cells, the differentiation of naive CD4+ T cells into “helper” T cells (Th1, Th2, Th17) and the plasticity among them is modulated; (4) the transcription of cytotoxic molecules (granzyme B and perforin) and activating receptors (NKG2D) is inhibited, and the expression of inhibitory KIR is enhanced in memory CD8+ T cells and NK cells, promoting allograft acceptance; (5) mature B cell differentiation into antibody-producing cells is controlled, decreasing allorecognition by donor-specific antibodies and preventing graft rejection. DNA methylation and demethylation are represented by black and white lollipops, respectively; histone modifications are shown as circles: green, H3K4me3; red, H3K27me3; purple, H3K9me3; blue, acetylation of H3 or H4.
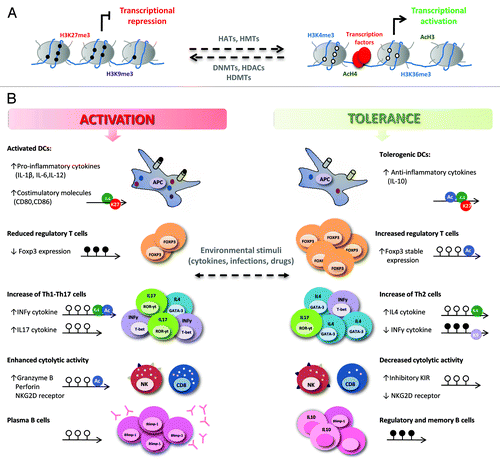
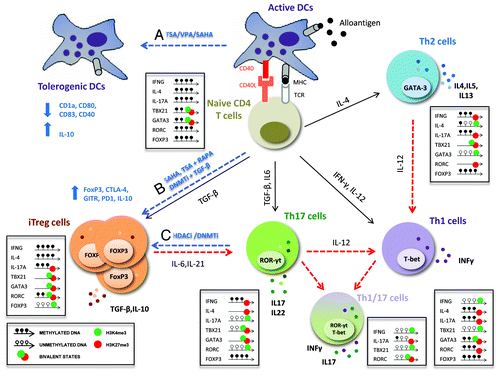
Figure 2. Targeting the activation and plasticity of CD4 T cells by HDAC inhibitors. After activation, CD4 T cells are directed toward different subsets of effector T cells (Th1, Th2 or Th17) or regulatory T cells (Treg) with specialized functions. These processes are regulated by epigenetic modifications that allow stable and heritable lineages but at the same time maintain the capacity to respond to environmental changes and switch from one lineage to another (plasticity). Dashed red lines indicate the plasticity and flexibility among CD4+ T cell subsets regulated by epigenetic mechanisms; dashed blue lines show the epigenetic treatments proposed for providing tolerance after transplantation. Epigenetic status of key transcription factors and cytokines essential for plasticity are shown for each CD4 T cell subset. This molecular mechanism may be related to poised, bivalent epigenetic stages (i.e., permissive H3K4me3 plus repressive H3K27me3 marks) in opposing lineages. HDAC inhibitors (HDACi) are believed to modulate the balance between immunity and tolerance: (A) TSA, VPA and SAHA diminish the expression of MHC class II and co-stimulatory molecules (CD1a, CD40, CD80, CD83), and disruption of HDAC11 increases IL-10 expression in DCs, favoring immune tolerance; (B) TSA and SAHA increase mRNA levels of FoxP3, CTLA4, GITR, PD-1 and IL-10, promoting the peripheral conversion of T cells into iTreg cells and enhancing suppressive function in vitro and in vivo; (C) an interesting approach is the use of epigenetic inhibitors to block the conversion of iTreg into Th17/Th1 cells in an inflammatory environment or the differentiation of effector T cells (Th1, Th17) into regulatory T cells with suppressive functions.