Figures & data
Figure 1 Redistribution of HP1α within the nucleus under heat shock conditions. (A) qRT-PCR analysis of hsp70 gene expression in control (untreated, C) and heat shock-treated (HS) MCF-7 cells. RNA extracted from treated and non-treated cells was reverse transcribed and the cDNA obtained was analyzed using a SYBR Green-based quantitative PCR approach. Amplification levels of hsp70 cDNA were normalized to the amplification level of GAPDH cDNA. Results of one representative experiment are shown. (B) Nuclear distribution of HP1α in control (untreated) and heat shock-treated cells. MCF-7 cells were immunostained with a mouse monoclonal antibody against HP1α and visualised by Alexa Fluor 488-conjugated anti-mouse IgG. DNA was stained with To-Pro 3 iodide fluorescent dye. Images were collected using a Leica laser scanning confocal microscope. Only one representative section is shown in each case. Bar scale: 5 µm. (C) Western blot analysis of HP1α in the nuclei (non-extracted and extracted with 0.5 M NaCl) of control (untreated, C) and treated with heat shock (HS) MCF-7 cells. Lamin B1 was used as a loading control.
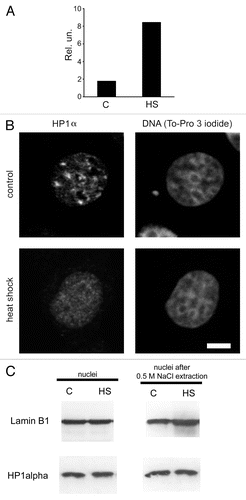
Figure 2 Redistribution of HP1α under heat shock conditions did not lead to decompaction of centromeric DNA. (A) Human MCF-7 cells, both untreated and heat shock-treated, were double immunostained with a human antibody against CENP-A (column 1) and a mouse monoclonal antibody against HP1α (column 2). Human and mouse primary antibodies were visualized by incubation with secondary antibodies conjugated to FITC and Alexa Fluor 555, respectively. Co-localization of CENP-A foci with HP1α is shown as yellow in the merged images (column 4). (B) Human MCF-7 cells, both untreated and heat shock-treated, were double immunostained with a human antibody against CENP-A (column 1) and a rabbit polyclonal antibody against histone H3 tri-methylated at lysine 9 (H3K9me3, column 2). Human and rabbit primary antibodies were visualized by incubation with secondary antibodies conjugated to FITC and Alexa Fluor 594, respectively. Co-localization of CENP-A foci with H3K9me3 is shown as yellow in the merged images (column 4). In (A and B) DNA was stained with DAPI fluorescent dye (column 3). Images were collected using a Zeiss LSM 510 META NLO multiphoton microscope. Only one representative section is shown in each case. Bar scale: 5 µm.
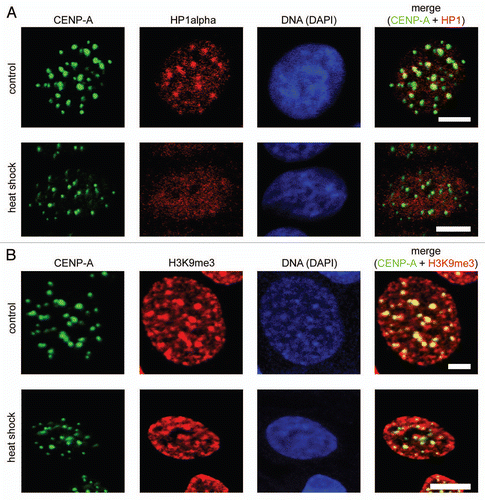
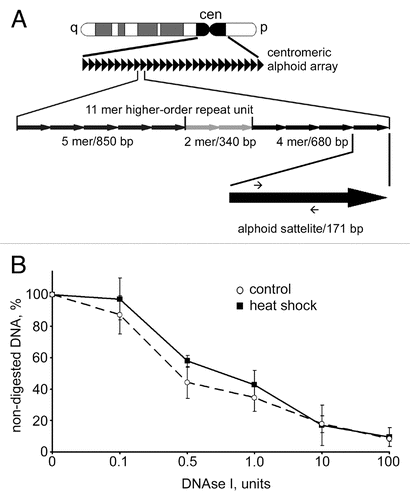
Figure 3 DNAse I sensitivity of centromeric DNA did not change upon heat shock. (A) A scheme representing the principal organization of centromeric alphoid arrays (for details see text). Positions of primers used for qPCR are depicted as small arrows on the black arrow that represents an individual 171 bp alphoid satellite. (B) Dynamics of digestion of alphoid satellite DNA by DNase I in untreated MCF-7 cells (dashed line) and heat shocked MCF cells (black line). Aliquots (50 ng) of genomic DNA extracted from nuclei of MCF-7 cells digested with increasing amounts of DNase I were subjected to SYBR Green-based quantitative PCR analysis. The Ct values obtained were converted to DNA concentration using a standard curve (data not shown). DNase I sensitivity was expressed as a percentage of preserved template for amplification of an alphoid satellite test fragment (y-axis) and is plotted for varying DNase I concentrations (0–100 U; x-axis). The data shown are an average of four independent experiments. Error bars represent the standard deviation for each concentration.