Figures & data
Figure 1. (A) Sonication times and fragments lengths in base pairs (bp) for cell line DNA at three different time periods: a:5 min; b:10 min; c:15 min; (B) Sonication times and fragments lengths in base pairs (bp) for fresh tissue DNA at three different time periods: a:5min; b:10min.
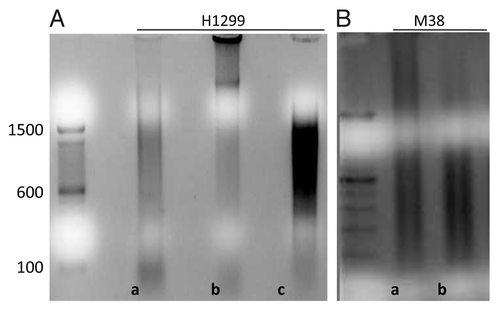
Figure 2. Agarose gel intensity after Nco I digestion for the two different aliquots of the same samples processed by three different MeDIP kits: Methylamp™ Methylated DNA Capture KIT (Epigentek), Methylated-DNA IP KIT (Zymo Research) and MagMeDIP KIT TM (Diagenode). T: Fresh Tissue DNA. C: Cell line DNA. MC: Methylated DNA Control. C+: Methylated/Non-methylated Control DNA
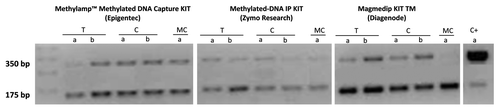
Table 1. Efficiency and consistency of the experiments performed in triplicate using two different amounts of initial DNA: 0.5ug and 1ug. DNA was isolated from fresh tissue (M38) cell lines (H1299) and fully methylated controls (C+) for the: MagMeDIP kit (Diagenode); Methylated-DNA IP Kit (Zymo Research); and MethylAmp™ Kit (Epigentek)
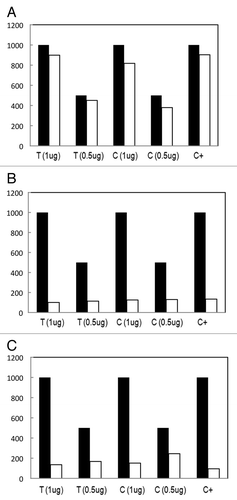
Figure 3. Reaction yield for different amounts of initial DNA, 0.5 μg and 1 μg of both, fresh tissue (T) and cell lines (C), for the: (A) MagMeDIP kit (Diagenode); (B) Methylated-DNA IP Kit (Zymo Research); and (C) Methylamp™ Kit (Epigentek).