Figures & data
Figure 6. Model of MAPK signaling. (A) Previous literature suggested that the subcellular localization of MAPK regulates the difference between growth and division and differentiation in developing fly wing and eye cells. (B) Modified model to incorporate additional regulation of growth and division by the phosphorylation status of the MAPK protein.
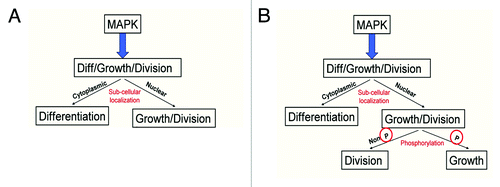
Figure 1A–F. Argos expression increases cell density while decreasing cell growth. (A-D) Third instar larval wing discs, anterior up, dorsal left, genotypes indicated bottom right, stain listed top right. (A) engrailed-Gal4; UAS:GFP (en::GFP) wing disc showing GFP expression (green) in the posterior compartment, and phosphorylated MAPK (pMAPK, red) throughout the wing pouch. (B) pMAPK staining (white) from (A). (C) engrailed-Gal4; UAS:GFP/UAS:Argos (en::argos) wing disc showing GFP expression (green) in the posterior compartment, and pMAPK (red) throughout the wing pouch. Note decreased pMAPK staining in the posterior compartment compared to the anterior compartment. (D) pMAPK staining (white) from (C). (E and F) Adult wings. (E) Normal wing venation pattern in engrailed-Gal4; UAS:GFP wing. (F) Phenotype of engrailed-Gal4; UAS:GFP/UAS:Argos adult wing. Note missing and decreased wing venation (arrow).
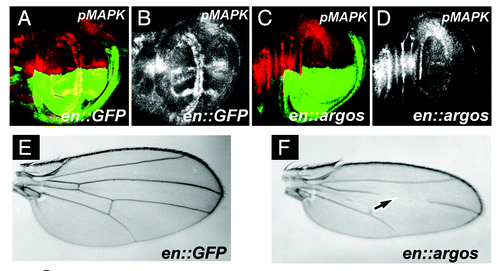
Figure 1G and H. Argos expression increases cell density while decreasing cell growth. (G) Cell Density in the posterior and anterior compartments of adult wings that express different proteins in their posterior compartment. Average cell density of the same area in the posterior and anterior compartments of each genotype is shown with Standard Error. All genotypes show engrailed:Gal4 expressing different transgenes, as indicated. (H) Surface area (expressed in pixel number) in the posterior and anterior compartments of adult wings that express different proteins in their posterior compartment. Average pixel number (representative of wing surface area) of the entire posterior and anterior compartments of each genotype is shown with Standard Error. All genotypes show engrailed:Gal4 expressing different transgenes, as indicated. In both graphs * indicates p < 0.05, while ** indicates p < 0.01.
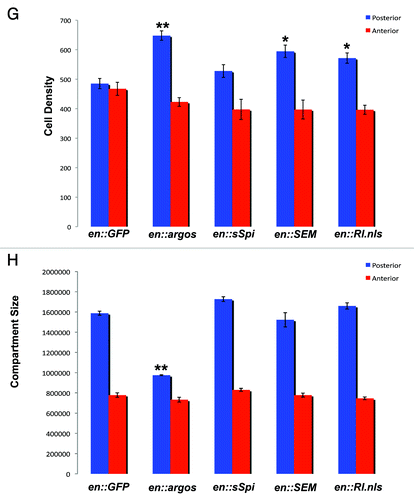
Figure 2. Increased pMAPK expression by sSpi expression is predominantly cytoplasmic. All panels show late third instar wing discs, anterior up, dorsal left, genotypes indicated bottom right, stain listed top right. (A) Wild-type wing discs shows GFP expression (green) in posterior compartment driven by the engrailed:Gal4 driver (en::GFP). Normal pMAPK expression (red) is indicated. (B) Expression of pMAPK (white) in engrailed-Gal4; UAS:RlSEM wing discs. Note increased expression of pMAPK in normal expression domains in posterior compartment. (C) Expression of pMAPK (white) in engrailed-Gal4; UAS:Rl.nls wing discs. Note no significant alteration of pMAPK expression compared to wild-type. (D) Expression of pMAPK (white) in engrailed-Gal4; UAS:sSpi wing discs. Note increased and expanded expression of pMAPK in posterior compartment. (E) Expression of pMAPK (red) and nuclear GFP (green) from red box in (D). Note that pMAPK is predominantly cytoplasmic. Scale bar indicates 5 μm. (F) pMAPK expression (white) from (E).
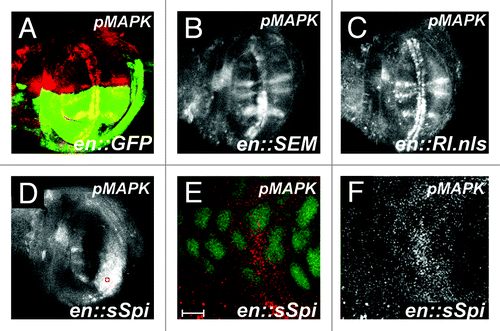
Figure 3. Argos expression does not decrease apoptosis in developing wings. All panels show wild-type third instar wing discs stained for activated Caspase 3. Anterior up, dorsal left. (A) Wild-type wing disc. (B) Positive control showing increased Caspase 3 activation (arrow) in the posterior compartment. Genotype is engrailed-Gal4/ UAS:DIM-7 (Drosophila Importin 7 homolog). (C) Caspase 3 staining (blue) in engrailed-Gal4; UAS:GFP/UAS:Argos discs. GFP is colored green. (D) Caspase 3 staining (white) from (C). Note there is no decrease in Caspase 3 activation upon Argos expression in the posterior compartment of wing discs.
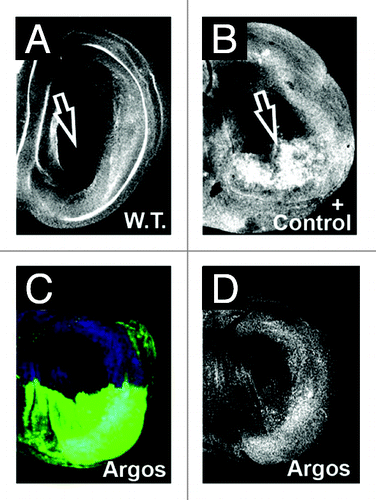
Figure 4. Phosphorylated MAPK, but not nuclear MAPK rescues Argos phenotype. (A) Shows ratio of posterior to anterior compartment of phosphorylated Histone H3 (pH3) staining in engrailed-Gal4; UAS:GFP third instar wing discs as compared to engrailed-Gal4; UAS:GFP/UAS:Argos and engrailed-Gal4; UAS:GFP/UAS:RlSEM wing discs. Note increased pH3 staining in engrailed-Gal4; UAS:GFP/UAS:Argos, indicative of increased mitotic events in the posterior compartment of these discs. (B) Cell Density in the posterior and anterior compartments of adult wings that express different proteins in their posterior compartment. Average cell density of the same area in the posterior and anterior compartments of each genotype is shown with Standard Error. Genotypes are indicated. (C) Surface area (expressed in pixel number) in the posterior and anterior compartments of adult wings that express different proteins in their posterior compartment. Average pixel number (representative of wing surface area) of the entire posterior and anterior compartments of each genotype is shown with Standard Error. Genotypes are indicated. In both graphs * indicates p < 0.05, while ** indicates p < 0.01.
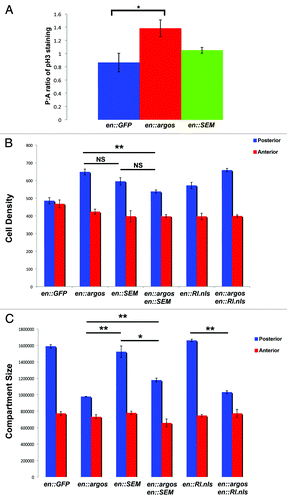
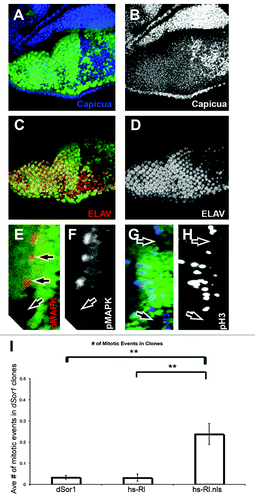
Figure 5. Nuclear, unphosphorylated MAPK increases cell division in Drosophila retinas. (A–H) Show third instar retinas, anterior up (A–D), anterior right (E–H). All retinas show clones of dSor1S-1221 marked by the absence of GFP (green). (A) Shows Capicua staining (blue) is increased in dSor1S-1221 clones. (B) Shows Capicua (white) from (A). (C) Shows ELAV (red, a marker of photoreceptor differentiation) is decreased in dSor1S-1221 clones. (D) Shows ELAV (white) from (C). (E) Shows phosphorylated MAPK (pMAPK, red) is absent in dSor1S-1221 clones. (F) Shows pMAPK (white) from (D). (G) Shows phosphorylated Histone H3 (pH3, blue) is absent from dSor1S-1221 clones in the second mitotic wave. (H) Shows pH3 (white) from (G). (I) Shows the average number of pH3 positive nuclei in dSor1S-1221 clones alone (first column), in dSor1S-1221 clones that also express full length MAPK within the clone (hs-Rl, second column), and in dSor1S-1221 clones that also express full length MAPK with a strong nuclear localization signal attached to it (hs-Rl.nls, third column). Error bars represent Standard Error. ** Indicates p < 0.01.