Figures & data
Figure 1. EHEC and EPEC OmpT degrade peptides at similar rates (A) Schematic of EHEC and EPEC ompT expressed from the croP promoter. Plasmids pEHompT and pEPompT are derived from pWSK129 and contain the C. rodentium croP promoter fused to the ompT open-reading frames of EHEC or EPEC, respectively. (B) Amounts of EHEC and EPEC OmpT produced by C. rodentium ΔcroP cells transformed with pEHompT or pEPompT were analyzed by western blotting using an antiserum developed against the CroP OM protease of C. rodentium (74% identical to OmpT).Citation14 (C) Cleavage of the synthetic FRET substrate containing the dibasic sequence RK in its center [2Abz-SLGRKIQI-K(Dnp)-NH2] was monitored over time by measuring fluorescence emission at 430 nm. (D) Proteolytic degradation of LL-37 by C. rodentium ΔcroP cells expressing EHEC or EPEC ompT. LL-37 was incubated for 1 h with the various bacterial strains. Aliquots were separated by Tris-Tricine SDS-PAGE (10–20% acrylamide) and gels were stained with Coomassie blue G-250. The control lane, in which no bacteria were added, contained 1.5 µg LL-37 in phosphate-buffered saline.
![Figure 1. EHEC and EPEC OmpT degrade peptides at similar rates (A) Schematic of EHEC and EPEC ompT expressed from the croP promoter. Plasmids pEHompT and pEPompT are derived from pWSK129 and contain the C. rodentium croP promoter fused to the ompT open-reading frames of EHEC or EPEC, respectively. (B) Amounts of EHEC and EPEC OmpT produced by C. rodentium ΔcroP cells transformed with pEHompT or pEPompT were analyzed by western blotting using an antiserum developed against the CroP OM protease of C. rodentium (74% identical to OmpT).Citation14 (C) Cleavage of the synthetic FRET substrate containing the dibasic sequence RK in its center [2Abz-SLGRKIQI-K(Dnp)-NH2] was monitored over time by measuring fluorescence emission at 430 nm. (D) Proteolytic degradation of LL-37 by C. rodentium ΔcroP cells expressing EHEC or EPEC ompT. LL-37 was incubated for 1 h with the various bacterial strains. Aliquots were separated by Tris-Tricine SDS-PAGE (10–20% acrylamide) and gels were stained with Coomassie blue G-250. The control lane, in which no bacteria were added, contained 1.5 µg LL-37 in phosphate-buffered saline.](/cms/asset/04d01263-827b-4280-a082-17cdf4498786/kgmi_a_10921656_f0001.gif)
Figure 2. OmpT protects EHEC from C18G-induced membrane disruption. EHEC wild-type and ΔompT strains were transformed with plasmid pCAST–ChFP encoding the OM localized red fluorescent protein mCherry.Citation20 Bacterial cultures (1ml) grown to mid-log phase were resuspended in 50 µl N-minimal medium and incubated for 15 min at room temperature in the absence or presence of the synthetic α-helical AMP C18G (100 µM). Bacterial cells (3 µl) were spotted on agarose-coated glass slides and analyzed under oil immersion at 100 magnification with a Zeiss Axiovert 200 M fluorescence microscope. (A) EHEC wild-type cell in the absence of C18G. (B) EHEC ΔompT cell in the absence of C18G. (C) EHEC wild-type cell in the presence of C18G. (D) EHEC ΔompT cell in the presence of C18G.
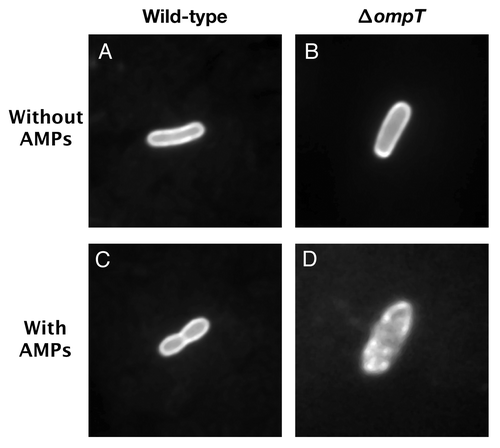
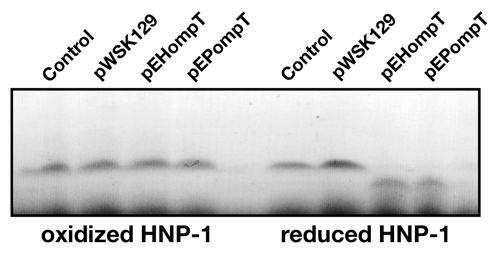
Figure 3. Cleavage of reduced HNP-1 by OmpT. HNP-1 was purchased from Peptides international Inc. (Louisville, KT). HNP-1 was reduced by incubation at 50°C for 30 min with dithiothreitol (1 mM). Free cysteines were then blocked with iodoacetamide (17 mM) for 30 min prior to incubation with bacterial strains. Oxidized or reduced HNP-1 was incubated for 1 h with the C. rodentium ΔcroP strain containing the empty plasmid pWSK129 or plasmid pWSK129 expressing EHEC OmpT (pEHompT) or EPEC OmpT (pEPompT). Bacterial cells were pelleted by centrifugation and supernatants were resolved on Tris-Tricine SDS-PAGE (10–20% acrylamide). Gels were stained with Coomassie blue G-250. Control lanes, in which no bacteria were added, contained 1 µg of oxidized or reduced HNP-1 in phosphate-buffered saline.