Figures & data
Figure 1. LPS transport from cytoplasm to periplasm in H. pylori. O-antigen assembly begins with the activity of WecA, which transfers an initiating GlcNAc-1-phosphate residue from an activated UDP donor to the lipid carrier undecaprenyl phosphate (UndP) at the cytosplasmic face of the inner membrane. The O-chain is extended by the alternating activities of galactosyltransferases and GlcNAc-transferases (purple), which receive their respective sugar substrates from a UDP-sugar donor. This generates a GlcNAc (green circles) and galactose (blue circles) sugar backbone to which fucosyltransferases A, B, and C transfer fucose residues (orange triangles) from an activated GDP-fucose donor. The Fut protein ruler mechanism is illustrated by length of protein corresponding to length of O-chain sugar that is fucosylated.Citation38 The completed O-chain containing Lewis antigens (patterned fucose residues linked to GlcNAc and galactose sugars) is translocated to the periplasmic face of the inner membrane by Wzk. Finally, it is removed from the UndPP carrier and transferred onto the lipid A/core by WaaL, generating a mature LPS species. UndPP is recycled to the cytoplasmic leaflet, releasing one phosphate group to reform the UndP acceptor.
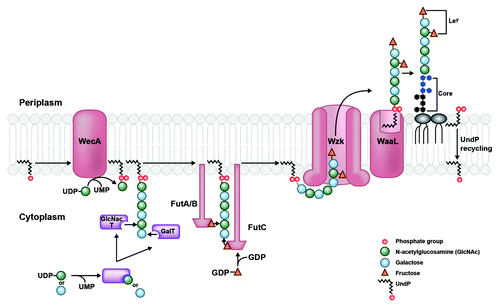
Figure 2. The Raetz pathway of lipid A biosynthesis in H. pylori. H. pylori synthesizes a bis-phosphorylated hexa-acylated lipid A via the conserved nine-step Raetz pathway using eight homologs of E. coli enzymes, and one enzyme (Hp0270) that is genetically distinct from its E. coli counterpart. The final product of this pathway, shown at the bottom left, is heavily modified during transport to the outer membrane.
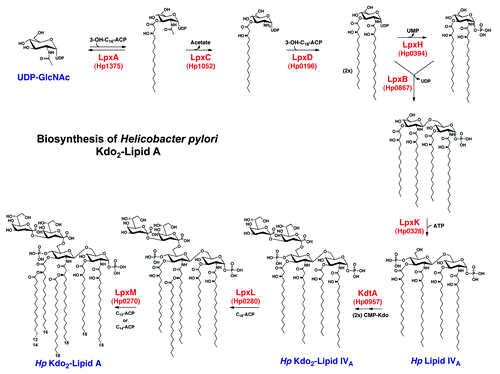
Figure 3. Modification of H. pylori lipid A generates its unique structure. Kdo2-lipid A is flipped to the periplasmic leaflet of the inner membrane by the conserved ABC-transporter MsbA. The 1-phosphate group is removed by LpxE (Hp0021), and a phosphorylethanolamine (pEtN) group is added in its place by EptA (Hp0022). The latter activity generates diacylglycerol (DAG), as phosphatidylethanolamine (PE) is likely the donor molecule for the pEtN group. Next, a two-protein Kdo-hydrolase complex, Kdo H1/H2 (encoded by hp0579 and hp0580), removes the terminal Kdo group. The last modification to occur before transport across the periplasm is the removal of the 4′-phosphate group by LpxF (Hp1580). Finally, the outer-membrane β-barrel LpxR (Hp0694) catalyzes removal of the 3′-linked acyl chains, generating the final lipid A structure in the outer membrane of H. pylori (inset). The core oligosaccharide and O-antigen chain with fucose additions are only shown in the inset for simplicity.
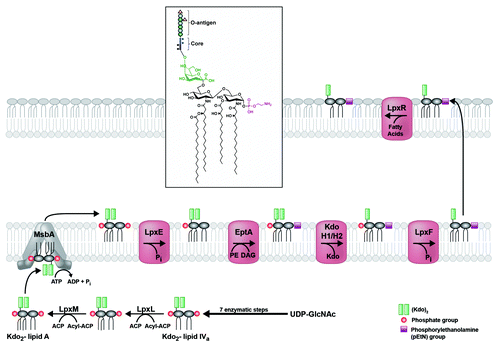
Figure 4. Lewis antigen structures. H. pylori can produce type 1 (left column) and type 2 (right column) Lewis antigens. (A) Type 1 Le antigens are based on a β(1,3)-linked galactose-GlcNAc sugar backbone, while type 2 antigens show a β(1,4) linkage. These backbones give rise to the Le antigens listed in panel (B). (C) Lea and Lex are built by α(1,4) or α(1,3) addition of a fucose residue to the GlcNAc sugar of the type 1 and type 2 backbone, respectively. (D) Leb and Ley are built by α(1,2) addition of a fucose residue to Lea and Lex structures, respectively.
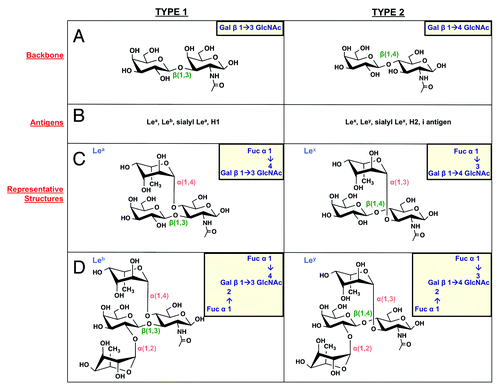
Figure 5.H. pylori peptidoglycan structure and modification. (A) The H. pylori PG monomer consists of an extended backbone chain of β(1,4)-linked GlcNAc-MurNAc sugars, linked to a short peptide chain by the 3-carboxyl group of the MurNAc residue. (B and C)H. pylori encodes a number of PG-modifying enzymes. Enzymes, genes, and activities are listed in table B, and the impact on the PG layer for each enzyme is illustrated in C.
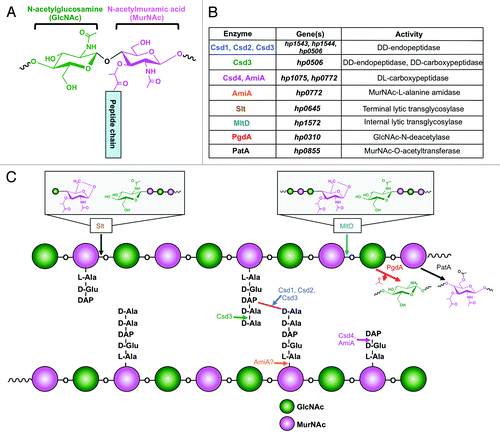
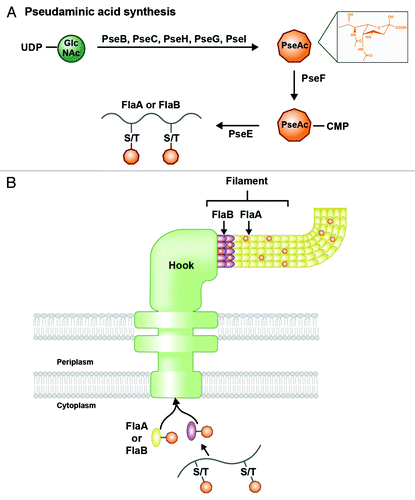
Figure 6. O-linked glycosylation of flagellar proteins in H. pylori. (A) Pseudaminic acid (PseAc) is synthesized in a five-step enzymatic process from a UDP-GlcNAc precursor. The structure of PseAc is shown (inset). It is then linked to a CMP nucleotide donor by PseF, and transferred by PseE from this donor to a serine or threonine residue on a FlaA or FlaB protein before the protein folds. (B) Glycosylated FlaA and FlaB proteins fold in the cytoplasm and display surface-exposed PseAc residues. Modified FlaA and FlaB are then translocated through the flagellar machinery to compose the flagellar filament. PseAc additions are apparent on both proteins within the filament.