Figures & data
Figure 1. Amino acid alignment of transmitted SIVmac251 Env sequences. (A) The identified SIVmac251 sequences and the reported SIVmac251 sequence CR2.RU.3R1 are compared with SIVmac239. (B) Cartoon depicts the location of the signal peptide, the variable (V) and conserved (C) regions, and gp41 of SIVmac239. The numbering follows SIVmac239 sequence. (C) Phylogenetic tree analysis of the SIVmac239 and the different SIVmac251 sequences. The tree also included 2 SIVsmE660 env CG7G and CG7V that are used in the neutralization assays.
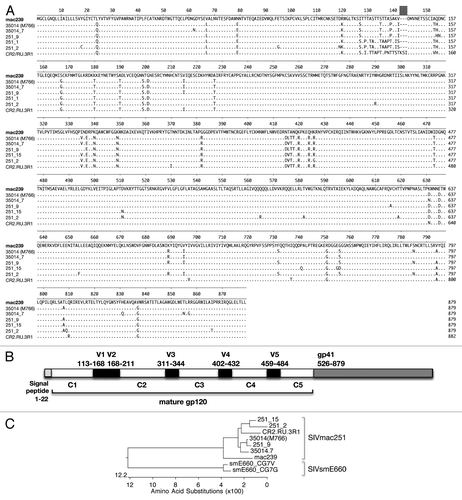
Figure 2. Induction of neutralizing antibody in macaques immunized via the intradermal (left panel) and the intramuscular (middle and right panels) route followed by EP. Neutralizing antibody titers were determined against SIVmac251-TCLA measured in M7-Luc cells (A) and against pseudotyped viruses containing the following Env: SIVmac251 35014 (M766) (B) and 35014_7 (C), the heterologous SIVsmE660 Env CG7G (D) and CG7V (E) were measured using the TZM-bl assay. The NAb measurements of a group of macaques which received 3 vaccinations of the same DNAs (total 2 mg) expressing the gp160 forms of Env via the IM/EP delivery is included (right panel). Neutralization titers shown are the log of the reciprocal dilution of sample that reduced the signal by 50% compared with virus in the absence of sample.
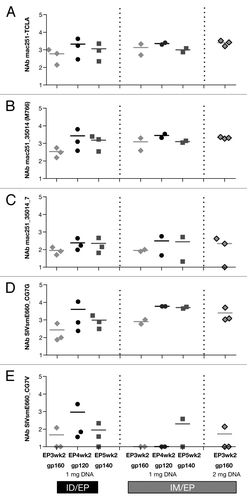
Table 1. SIV Env and functional characteristics
Figure 3. Characterization of different SIV mac251 Env proteins. (A) Western immunoblot analysis of the expression of SIVmac239 Env and the different SIVmac251 Envs upon transient transfection of HEK293 cells. (B) Cartoon depicting gp160, gp140 and gp120 Env. (C) Western immunoblot analysis the gp160, gp140 and gp120 Env of SIVmac251_35014 (M766) upon transient transfection of HEK293 cells. Cell-associated and extra-cellular fractions are shown. (D) Pseudotyped recombinant HIV-luc virus reporter (NL4–3.LucR−E−) viruses were generated using the SIVmac239 Env and the indicated SIVmac251 Env. Infectivity was tested in modified CEM-X174 cells that contain the HIV-LTR promoter linked to GFP stably integrated. GFP expression, which serves as read-out for infection, was measured by FACS analysis.
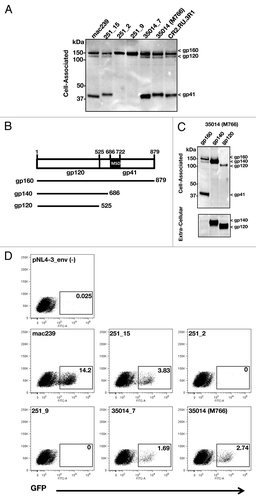
Figure 4. Immunogenicity of Env DNAs in mice. (A) Outline of vaccination study of BALB/c mice. Mice (n = 5/group) were vaccinated via the IM route (needle and syringe) at 0 and 4 weeks with SIV Env DNA formulated in PBS, and were sacrificed 2 weeks after the 2nd vaccination. Spleen and blood were analyzed. (B) Splenocytes were stimulated with an Env peptide pool (15-mer, 11 AA overlap) and the IFN-γ producing T cells were measured by ELISPOT assay. (C) The Env-specific binding antibodies titers were measured by ELISA using serial dilutions of pooled plasma samples.
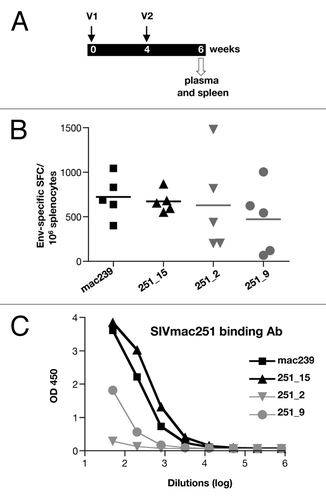
Figure 5. Production and purification of SIV Env protein from stable HEK293 cells. (A) Samples of culture media from Fibercell grown HEK293 cells expressing SIV env protein were assessed for purity on Coomassie blue stained SDS PAGE gels (10% TGX gel, Bio-Rad). (B) Purified SIVsmE660_CG7V gp140, SIVmac gp140 (35014_7) and gp120 (35014/M766) proteins were assessed for purity and quality on non-denaturing gel (10% TGX gel) and visualized by Coomassie blue staining.
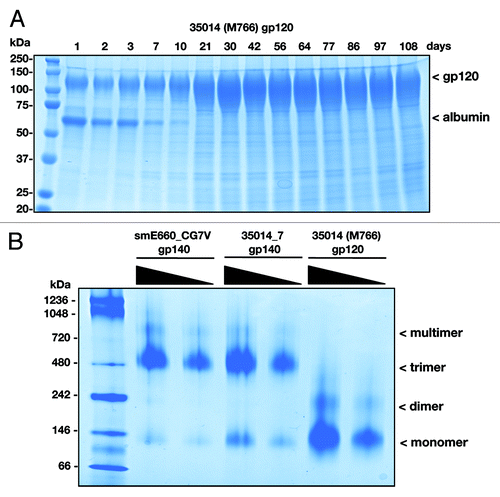
Figure 6. Analysis of humoral immune responses upon vaccination of macaques with env DNA. (A) Outline of vaccination study of macaques indicating the sequential immunization with DNA mixtures expressing a cocktail of multivalent Env gp160 (EP1-EP3), gp120 (EP4) and gp140 (EP5). (B) Endpoint SIVmac251 Env binding Ab titers are shown for the animals vaccinated ID/EP and the IM/EP DNA delivery regimen using the same DNA mixtures.
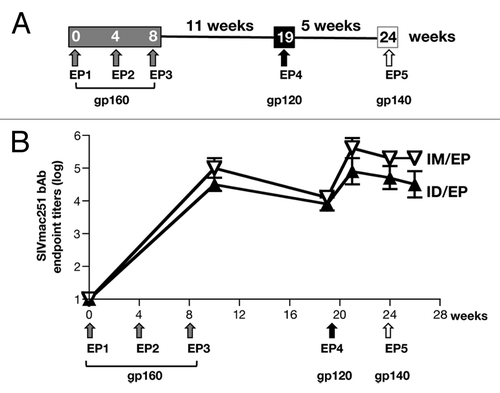
Figure 7. Persistence of humoral responses in macaques vaccinated via the intradermal and the intramuscular route. The animals were monitored for 14–20 mo after the last vaccination. Humoral immune responses were measured in the macaques that received DNA via the intradermal (n = 3) and the intramuscular route (n = 5, with 3 macaques receiving 1 mg DNA and 2 macaques receiving 2 mg DNA). (A) Endpoint binding Ab titers to SIVmac239, (B) NAb titers to SIVmac_35014 (M766) and (C) NAb to the heterologous SIVsmE660_CG7G (tier 1A-like) are shown as log of reciprocal dilution of plasma that produces a 50% reduction in signal compared with wells receiving no sample.
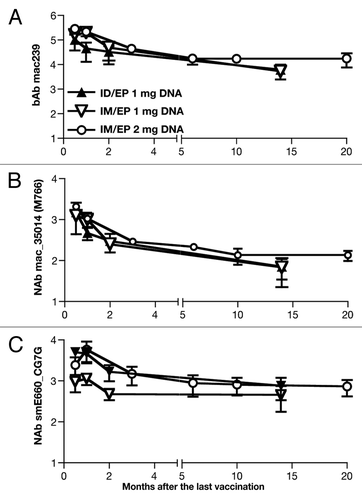
Table 2. Avidity of SIVmac239 Env binding antibody over time
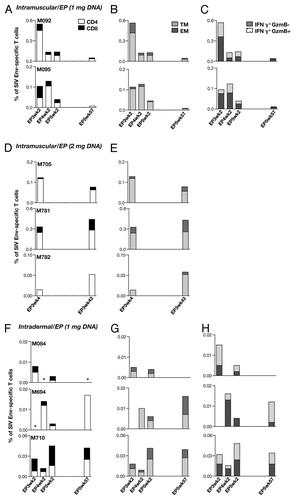
Figure 8. Comparison of magnitude and longevity of cellular responses in macaques vaccinated via the intramuscular (A-C, D-E) and the intradermal (F-H) route. Cellular immune responses were measured in PBMC upon stimulation with a SIVmac239-specific peptide pool (15-mer overlapping by 11 AA) at 2 weeks after vaccination with gp160 (EP3wk2), gp120 (EP4wk2) and gp140 (EP5wk2) (A-C, F-H). Animals in D-E were monitored after the 3rd gp160 DNA vaccinations. All animals were monitored for ~1 y and the cellular responses were measured at EP5wk57 and EP3wk43, respectively. The responses are shown as % Env-specific IFN-γ+ T cells among CD4+ and CD8+ (A, D, F); transitional (TM; CD28+CD95+CCR7-) and effector memory (EM; CD28-CD95+CCR7-) (B, E, G) T cells. The frequency of the Env-specific IFN-γ+ granzyme B+ T cells is shown (C, H). The macaques of panels B and E were not analyzed for this function. Asterisk indicates that the sample scored negative for the assay.