Figures & data
Figure 1. IRR visualizes blood flow in the non-fluorescent sinusoidal microvasculature. (A) IRR was obtained with a 633 nm HeNe laser at 25% power output and collected at 630–850 nm. In this 3D projection, sinusoids appear as red tubes due to the sinusoidal wall and dense packing of blood cells reflecting the excitation light. (B) ECFP mouse liver fluorescence was excited with a 405 nm Diode laser to visualize the hepatocyte plates. (C) Overlay shows the intricate network of liver parenchyma and highly anastomozed sinusoids. (D) Inverse IRR highlights the sinusoidal microvasculature, while DsRed mouse fluorescence, obtained with 543 nm HeNe laser, visualizes the hepatocyte plates (E). (F) Overlay allows for a 3D representation of the non-fluorescent structural details of the sinusoids (red) and the DsRed fluorescence of the liver tissue (green pseudocolor). Scale bars 20 μm.
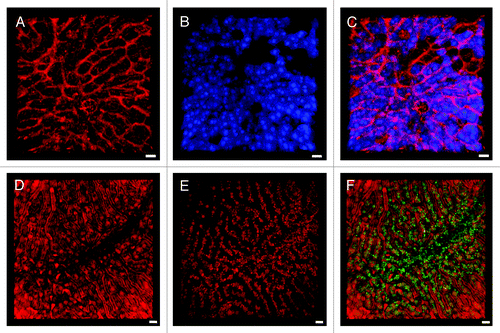
Figure 2. Cell membrane and mitochondrial staining visualizes the sinusoidal liver architecture. (A–C) In the liver, the cell membrane stain CellMask Orange (shown in green) highlights the sinusoidal microvasculature due to the high density of cell membranes along the space of Disse. (A) 3D reconstruction of a z-stack. (B) A z-slice from the avascular subcapsular region of the liver demonstrates that CellMask Orange predominantly outlines hepatocytes. (C) Another z-slice, acquired 40 μm from the liver surface, visualizes that CellMask Orange outlines the microvasculature as well as individual blood cells within the sinusoidal lumen. (D–F) Liver from a mouse injected with MitoTracker Deep Red (D) and CellMask Orange (E). The overlay (F) visualizes the sinusoidal microvasculature outlined with CellMask Orange, while the hepatocyte cytoplasm is filled with red fluorescent mitochondria. Distortion of the RBCs in the sinusoidal lumen indicates preserved blood flow. Scale bar 20 μm.
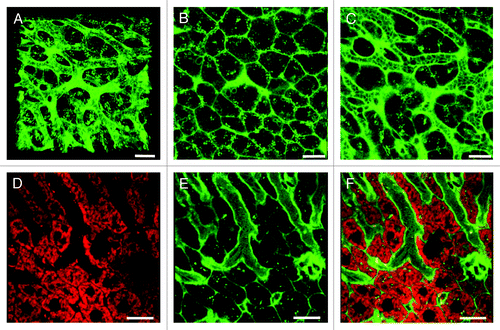
Figure 3. Novel complementary approaches for visualization of the sinusoidal microvasculature. (A) In this 3D reconstruction of a lysM-EGFP mouse liver z-stack, the sinusoidal microvasculature is labeled with anti-CD31 Alexa Fluor 647 (red). Myelomonocytic cells in this reporter mouse fluoresce green and nuclei are blue (Hoechst). (B) Z-slice of a Tie2-GFP mouse liver with fluorescent endothelia (green). Hepatocytes are red (MitoTracker), CD8+ T cells were labeled with anti-mouse CD8a-PE (green outline) and nuclei are blue (Hoechst). See Vid. S8 for complete z-stack. (C) 3D projection of a liver z-stack showing anti-CD8a-PE labeled CD8+ T cells (red circles) patrolling the sinusoidal endothelium (green). Nuclei are blue (Hoechst).(D) Z-slice showing red hepatocyte plates (MitoTracker) lined with green fluorescent sinusoidal endothelia. Nuclei are blue (Hoechst). Scale bars 20 μm.
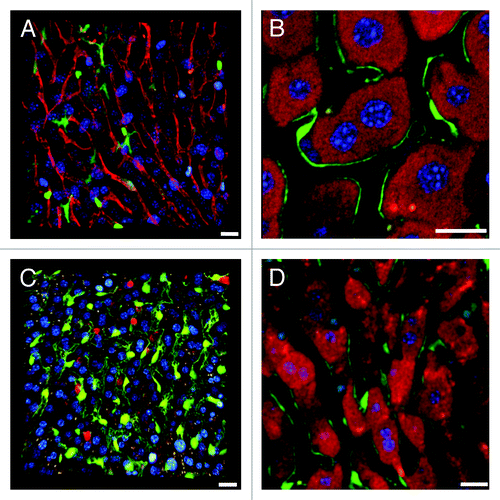
Figure 4. Complementary imaging techniques for reporter mice with fluorescent immune cells. (A) Single z-slice of a lysM-EGFP mouse liver with fluorescent myelomonocytic cells (solid green cytoplasm, in this case a Kupffer cell). MitoTracker was used to visualize hepatocytes (red), CD8+ T cells were labeled with anti-mouse CD8a-PE (green outline) and nuclei are blue (Hoechst). (B) 3D reconstruction of a z-stack from a CD11c-EYFP mouse liver with fluorescent dendritic cells (yellow). Parenchymal cells are red (MitoTracker), nuclei are blue (Hoechst). Scale bar 50 μm. (C) A DsRed x lysM-EGFP hybrid mouse has red fluorescent hepatocytes and green fluorescent Kupffer and myelomonocytic cells. All other scale bars 20 µm.
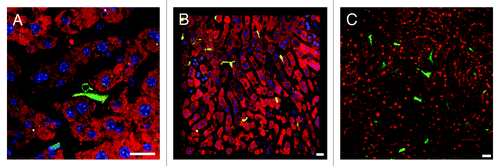
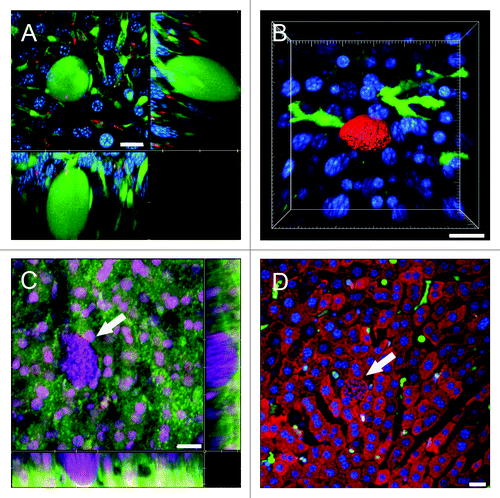
Figure 5. Using IRR to visualize the sinusoidal blood flow surrounding Plasmodium LS in non-fluorescent mice. (A) 3D volume view of a mouse liver harboring a 22 h PyXNL-GFP LS (green). Hepatocytes are labeled with MitoTracker (red), nuclei are blue (Hoechst). (B) MitoTracker (red) labels the hepatocyte plates surrounding two 42 h PyXNL-GFP LS (green). (C) IRR highlights the sinusoidal microvasculature (red) surrounding a 42 h PyXNL-GFP LS (green). Optical xy-xz-yz slice view, obtained with 50% 594 nm and 633 nm HeNe laser output power and PMT set to 590–850 nm. Nuclei are blue (Hoechst). (D) 3D volume view shows a 42 h PyXNL-GFP LS (green) embedded in non-fluorescent liver tissue. The dense sinusoidal network (red) was visualized using 50% 594 nm and 633 nm HeNe laser output power and setting the PMT to 590–850 nm. Nuclei are blue (Hoechst). Scale bars 20 μm.
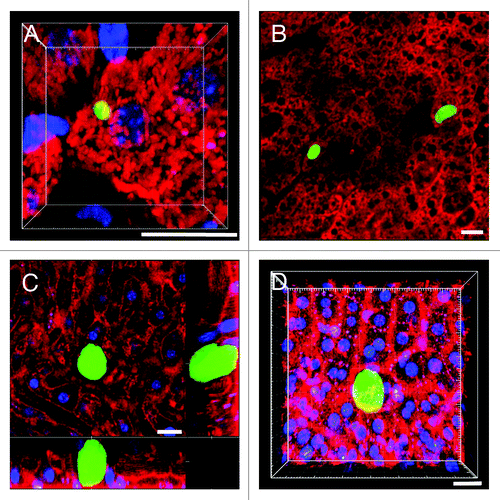
Figure 6. Visualization of the sinusoidal architecture aids in analysis of the Plasmodium infected liver. (A) Optical xy-xz-yz slice view of a 42 h PyXNL-GFP LS in a Tie2-GFP mouse liver. Despite expression of the same green fluorescent protein, endothelia can readily be distinguished from the large LS (arrows). Kupffer cells were identified by phagocytic uptake of dextran-rhodamine (red). (B) 3D reconstruction showing a 42 h PyXNL-DsRed LS (red) surrounded by dendritic cells (green) in a CD11c-EYFP mouse liver. Nuclei are blue (Hoechst). (C) Optical xy-xz-yz slice view of a non-fluorescent 42 h PyXNL-wt LS in a DsRed mouse liver. The parasite is identifiable by the large number of small blue merozoite nuclei (Hoechst). The sinusoidal architecture was visualized by IRR (green) using 40% output power of a 633 nm HeNe laser and PMT set to 630–750 nm. DsRed fluorescence is most pronounced in hepatocytes. See Figure S2 for individual color channels. (D) A non-fluorescent 42 h PyXNL-wt LS in a lysM-EGFP mouse with green Kupffer and myelomonocytic cells. Hepatocytes are red (MitoTracker). The small punctate merozoite nuclei of the mature LS can readily be distinguished from the much larger hepatocyte nuclei (both stained blue with Hoechst). Scale bars 20 µm.