Figures & data
Table 1. Characteristics of the antibodies in the affinity maturation sequence analysis data set
Figure 1. Summary of the amino acid bias observed during the in vitro affinity maturation of 36 individual CDR loops from 21 different monoclonal antibodies (mAbs). (A) shows the range of potency gains for each of the 21 mAbs upon which affinity maturation was performed. The 21 mAbs are grouped according to the antigens they were selected on and data points colored gray or white according to antigen grouping. The affinity matured variant sequences were subsequently included in an analysis for amino acid bias, which is summarized in (B–D). (B) shows the frequency, in the overall data set of VH and VL CDR3 loops, with which each amino acid type was categorised as "selected for" (black bars), "neutral" (light gray bars) or "selected against" (white bars). Amino acids are grouped in six families from left to right, neutral and small, special, polar and relatively small, polar and relatively large, nonpolar and relatively small and nonpolar and relatively large. The representative amino acids from five of the six families that were prioritized for further study on the basis of being "selected for" during the affinity maturation process are highlighted with asterisks. In (C and D), the equivalent data are plotted for VH CDR3 loops alone and VL CDR3 loops alone, respectively.
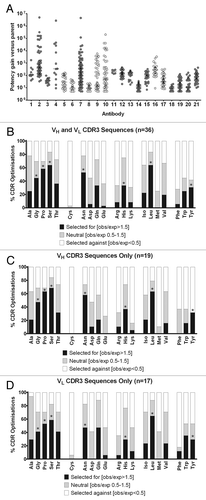
Figure 2. Representation of the CDR loop randomization strategies for affinity maturation. In the typical in vitro optimization strategy, blocks of six CDR residues are fully randomized in separate libraries using degenerate NNS codons, producing the amino acid frequencies shown in the graph. In contrast, the TiAM approach uses an equal frequency of eight different codons (encoding the wild type plus the seven preferred amino acid sub-types), allowing up to 12 residues to be mutated in parallel in a single library. Amino acids are grouped in six families according to side-chain properties, neutral and small, special, polar and relatively small, polar and relatively large, nonpolar and relatively small and nonpolar and relatively large.
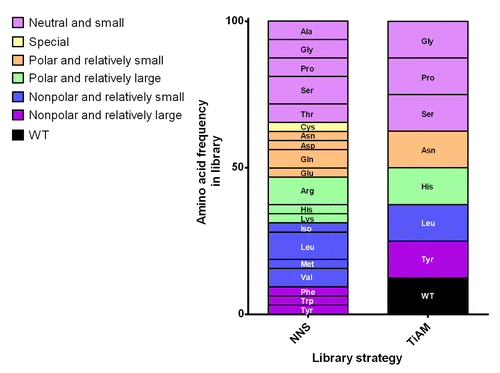
Figure 3. Comparison of affinity gains from the standard NNS optimization approach and the TiAM method. (A) The affinity improvement over the ICM10064 parent antibody for all optimized VLCDR3 variants from the TiAM and NNS methods, respectively. (B) The affinity improvement over the ICM10088 parent antibody for all optimized VHCDR3 and VLCDR3 variants from the TiAM and NNS methods. Also plotted are the improvements conferred by performing the TiAM approach in VLCDR 1 and 2 and VHCDR 1 and 2.
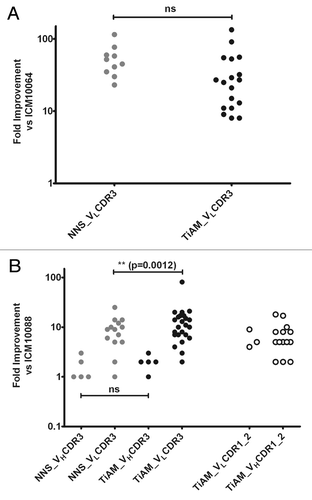
Figure 5. Sequence summary for variants which have undergone TiAM and NNS optimization of VLCDR3 loops in antibodies ICM10064 and ICM10088. For lineage ICM10064 (A) and ICM10088 (B), the wild type VLCDR3 loop sequence is shown at the top and for each variant the changes incorporated during the affinity maturation process are highlighted. Amino acids are colored according to side-chain properties, as detailed in the legend, and the improvement in affinity measured for each individual variant is recorded.
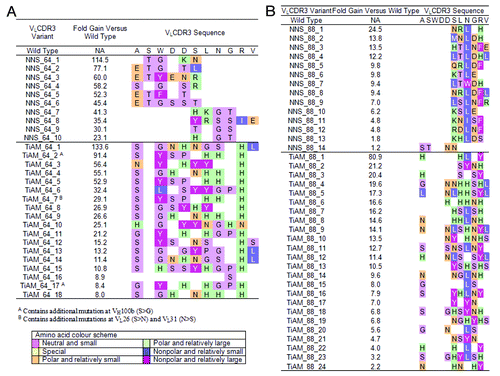
Figure 4. Dose titration analysis of lead antibodies comparing the affinity gains of antibodies from the biased library strategy vs. the standard optimization approach. For lineage ICM10064 (A), the most potent scFv antibodies from the NNS and TiAM approaches respectively, were NNS_64_1 and TiAM_64_1. In the ICM10088 lineage (B), the most potent scFv antibodies from the NNS and TiAM approaches, respectively, were NNS_88_1 and TiAM_88_1. Full dose titration curves, with standard error bars, are plotted and calculated IC50 values are shown for all antibodies.
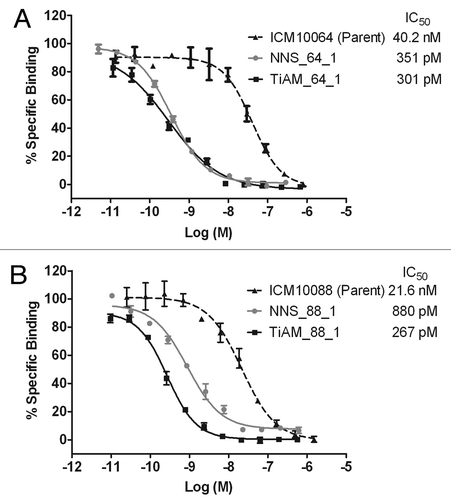
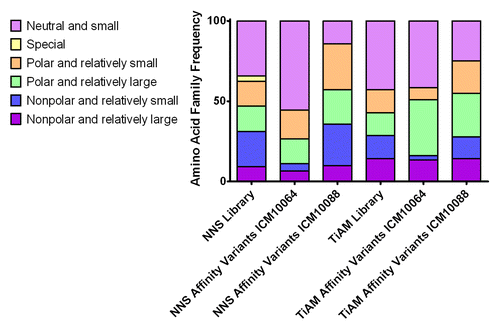
Figure 6. Amino acid frequency in TiAM and NNS libraries before and after selection for improved affinity. The amino acid distribution before library selection was calculated based on the respective library designs and the distribution after affinity selection was calculated from the CDR loop sequences of the affinity matured ICM10064 and ICM10088 variants. Amino acid families are colored according to the key.