Figures & data
Table 1. Summary of anti-HB-EGF mAb generation by mouse strain and immunogen
Figure 1. Neutralizing activity of anti-HB-EGF mAbs against sHB-EGF functions. (A) Neutralizing activity of anti-HB-EGF mAbs against sHB-EGF-induced EGFR phosphorylation in SK-OV-3 cells. The plots represent the IC50 values of 146 mAbs used in the epitope binning study (). The horizontal line indicates the median IC50 value. (B) Neutralizing activity of anti-HB-EGF mAbs against sHB-EGF-induced colony formation in RMG-I cells. We used the 89 mAbs with IC50 values of less than 5 × 10−9 M in the EGFR phosphorylation assay, and this plot represents the IC50 values of the 83 mAbs with IC50 values of less than 1 × 10−7 M. The horizontal line indicates the median IC50 value. (C) Correlation of neutralizing activities of anti-HB-EGF mAbs against sHB-EGF-induced EGFR phosphorylation and colony formation. Each dot represents the IC50 values of 83 mAbs with an IC50 value of less than 1 × 10−7 M in the colony formation assay (B). The data were analyzed by the Spearman’s test (two-tailed). The Spearman correlation was r = 0.6730 (p < 0.0001).
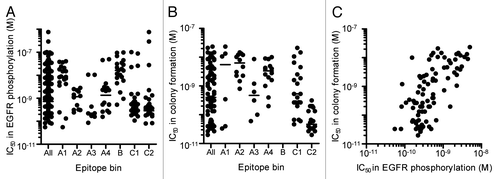
Figure 2. Binding activities of neutralizing anti-HB-EGF mAbs to HB-EGF. Binding activity of anti-HB-EGF mAbs to (A) human sHB-EGF, (B) mouse sHB-EGF and (C) rat sHB-EGF. The binding activities were determined by ELISA. The plots represent EC50 values of 146 mAbs used in the epitope binning study (). The horizontal line indicates the median EC50 value. (D) Binding activity of anti-HB-EGF mAbs to proHB-EGF was measured by flow cytometry. The plots represent median fluorescence intensity (MFI) ratio of the 146 mAbs used in the epitope binning study (). The MFI ratio of anti-HB-EGF mAb to a control sample was used as the measure of binding activity to proHB-EGF. The horizontal line indicates the median MFI ratio.
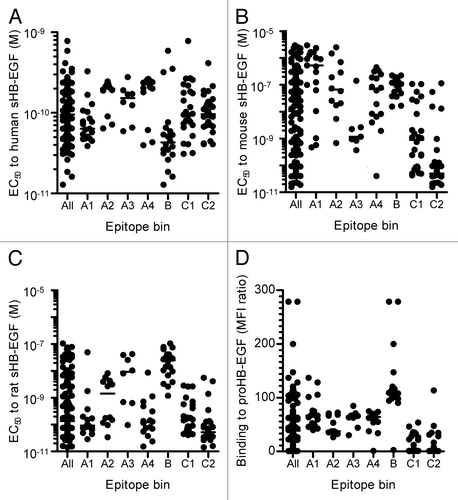
Figure 3. Epitope binning study of anti-HB-EGF mAbs. The competitive binding of all mAb pairs to sHB-EGF was measured in the FMAT assay. Binding of biotinylated anti-HB-EGF mAb to sHB-EGF was detected in the presence of a label-free anti-HB-EGF mAb. Horizontal and vertical axes indicate unlabeled and biotinylated mAbs, respectively. Data were analyzed by Ward’s method with Euclidean distance. Dendrogram on the left axis represents the similarity of each mAb in the competitive binding pattern to sHB-EGF. The color scale from 0 to 100 shows the competitive binding of the 2 mAbs.
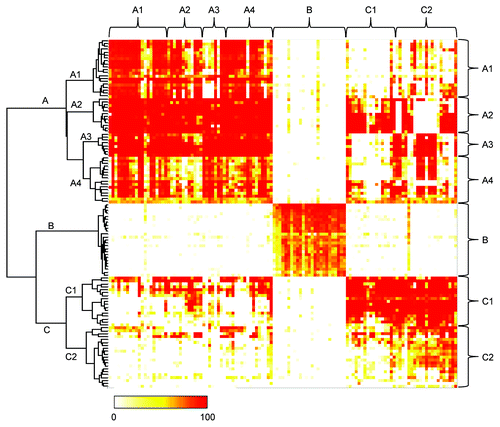
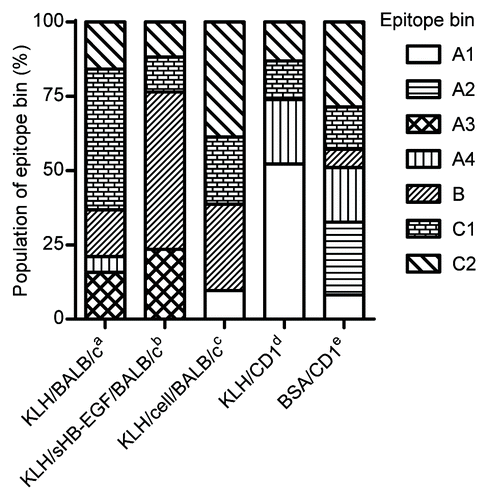
Figure 4. Difference of epitope bin population by mouse strain and immunogen type. Epitope bin populations were analyzed based on the immunization method used to generate the mAbs. a-cBALB/c immunization with aKLH-conjugated sHB-EGF; bKLH-conjugate and a final boost of sHB-EGF; cco-immunization of the KLH-conjugate and proHB-EGF-expressing cells; d,eCD1 immunization with dKLH-conjugate; eBSA-conjugated sHB-EGF.