Figures & data
Figure 1. Expression and purification of bispecific TandAb antibodies, recombinant CD3 antigen and localization of EDB in murine tumors. (A) Schematic representation of bispecific antibodies in the IgG and diabody formats. (B) Bispecific antibodies mCD3εxEDB and HELxEDB in the TandAb format used in this study. (C) SDS PAGE of purified TandAbs under non-reducing conditions showing migration at the expected (monomeric) size of 55 kD. (D) Size exclusion profile (Sephadex 200) of purified TandAb dimer after an initial polishing step (data not shown) showing a single peak corresponding to the expected size of 110 kD. (E) Schematic representation of the CD3 antigen (mCD3εγ26) used in this study, Coomassie staining and anti-His western blot of the purified antigen. (F) Size exclusion chromatography of purified mCD3εγ26. (G) Sections of F9 teratocarcinoma tumors excised from 129Sv mice. The first and third panels show immunohistochemical staining of blood vessel endothelial marker CD31 (green). The second panel shows immunohistochemical staining of tumor antigen EDB (red). The fourth panel shows staining of the tumor with antibody against the irrelevant hen egg lysozyme antigen, as negative control. For detection, biotinylated L19 small immunoprotein (SIP), revealed with Alexa594-conjugated streptavidin (Invitrogen) (red), was used. Biotinylated KSF SIP (binding the irrelevant hen egg lysozyme antigen) was used as negative control.
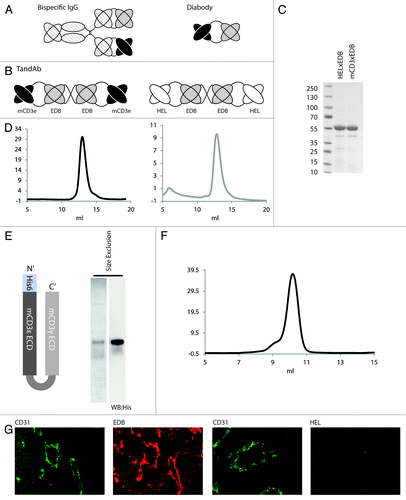
Figure 2. Binding affinity of TandAbs to CD3 and EDB antigens in vitro. (A) Binding of mCD3εxEDB to immobilized mCD3εγ26, and (B) to EDB at indicated concentrations of antibody. (C) Binding of the parental 145-2c11 IgG antibody to mCD3εγ26. (D) Binding profile of negative control HELxEDB antibody to mCD3εγ26. (E) FACS of CTLL2 cells stained with mCD3εxEDB and protein A-Alexa488 conjugate (black) or with Protein A-Alexa488 conjugate alone (gray). (F) Cells stained with 60 ng mCD3εxEDB (black) or HELxEDB (white) and revealed with protein A-Alexa488 conjugate. (G) Cells stained with mCD3εxEDB and EDB-FITC conjugate (black line) or EDB-FITC conjugate alone (gray line).
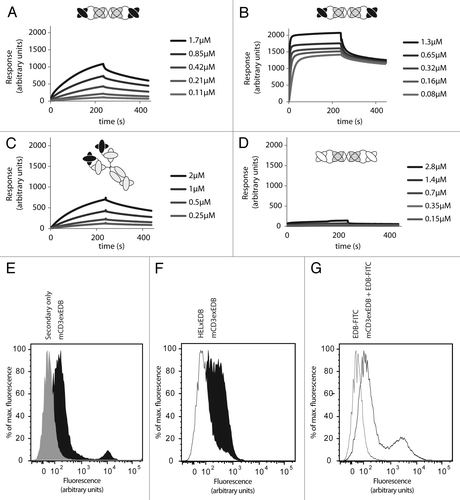
Figure 3. Biodistribution of 125I labeled TandAbs in immunocompetent mice bearing syngeneic tumors. Immunocompetent mice bearing F9 syngeneic tumors were injected with 10 µg (0.09 nmol) 125I radiolabelled (A) mCD3εxEDB TandAb and (B) HELxEDB TandAb. Bars show the mean ± SE percentage injected dose per gram of tissue (%ID/g). Bioreactivity after radioiodination was confirmed by binding to EDB resin (data not shown).
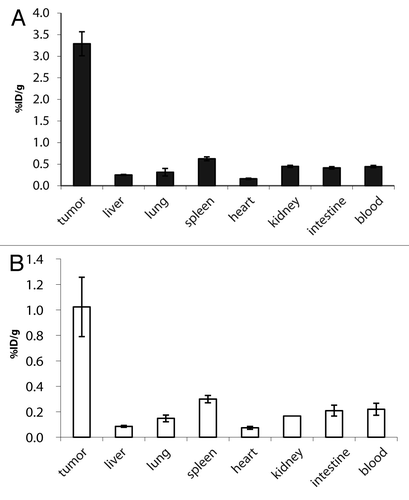
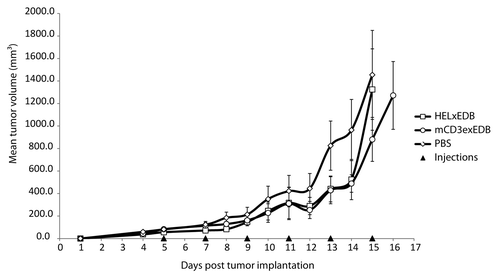
Figure 4. Therapy with mCD3εxEDB TandAb in mice. Mean tumor volume ± SD (mm3) is shown over time in 129Sv mice (n = 6 per treatment group) implanted s.c. with F9 tumor cells. Mice were injected i.v. in the tail vein with 40 µg TandAb mCD3εxEDB, HELxEDB or saline solution every 48 h for 6 injections in total.