Figures & data
Figure 1. Affinity determination of the CRP specific scFv antibodies by SPR analysis. CRP was covalently coupled to a CM5 sensor chip. Sensorgrams of the CRP specific scFv antibody clones LA13-IIE3 (A), LA13-IIC3 (B) and LA13-IID4 (C) are shown. Four to five scFv concentrations were used to determine the affinity constants KD and off rates koff (D).
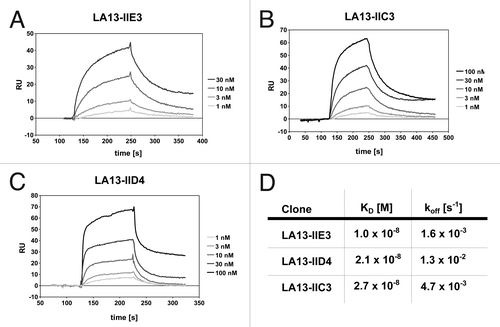
Figure 2. Size-exclusion chromatography of affinity-purified scFv LA13-IIE3 on a calibrated Superdex 200 10/300 GL column. Molar mass calibration (closed orange squares) was done with blue dextran (2,000 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa) and cytochrome C (12.4 kDa).
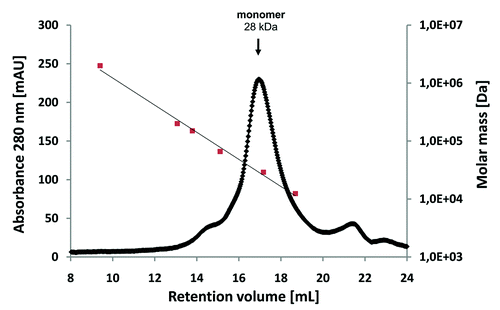
Figure 3. Epitope binning analysis of the CRP-specific scFv antibodies using SPR analysis. Overlay plots of SPR sensorgrams are arranged according to the scFv antibody injected first: LA13-IIE3 (A), LA13-IIC3 (B), and LA13-IID4 (C). All scFv antibodies were injected with saturating concentrations according to the CRP coupled CM5 sensor chip. Flow rate were kept constant throughout the measurement. The second injection of a different scFv antibody than the first one (run 4, 5, 9, 10, 14, and 15; red or blue solid lines) resulted only in an increased response signal if antigen binding of first and second scFv antibody did not interfere with each other. Injection of saturating concentrations of solely the first (runs 2, 7, and 12; gray dotted lines) or solely the second (runs 1, 6, and 11; black dashed lines) scFv antibody resulted into a response curve with typical dissociation and association pattern. Continuous injection of the first scFv antibody (runs 3, 8 and 13; green solid lines) were included as additional controls.
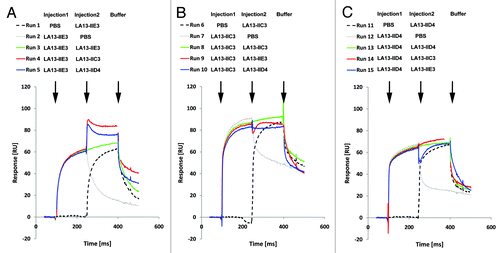
Figure 4. Immunoblot of CRP using scFv antibodies LA13-IIE3, LA13-IIC3 and LA13-IID4. CRP was prepared in SDS sample buffer (5 min, 98°C). A total of 150 ng CRP per lane was electrophoretically separated in a 12% (w/v) SDS polyacrylamide gel and transferred onto PVDF membrane. Immunostaining was performed with the CRP specific scFv antibodies LA13-IIE3, LA13-IID4, and LA13-IIC3 followed by the myc-tag specific mAb Myc1-9E10 and an AP conjugated secondary antibody conjugate. BSA was used as negative control.
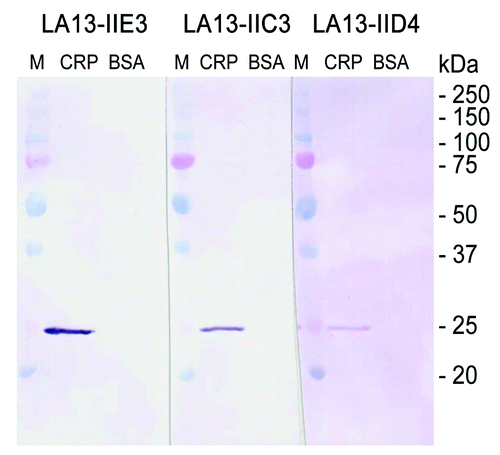
Figure 5. Epitope mapping of CRP specific scFv LA13-IIC3, LA13-IID4, LA13-IIE3. A series of overlapping 15mer oligopeptides covering the whole CRP protein sequence with an offset of three amino acids was synthesized onto filter membranes. These filter membranes were subsequently immunostained with CRP specific scFv antibodies LA13-IIC3 (A), LA13-IID4 (B), and LA13-IIE3 (C). Bound scFv antibodies were detected with α-myc-tag antibody and secondary antibody AP conjugate. Both epitopes are highlighted (yellow, space fill) in one subunit of the CRP pentamer structure (D). Epitope location based on structure (PDB ID: 1B09)Citation4: peptide backbone (gray ribbon), side chains (lines), and polyhendron surface (points) are shown (visualization with 3D-Mol Viewer), epitopes indicated in yellow. Important amino acid residues of the epitopes identified by an additional alanine walk (data not shown) are underlined (* only IIC3)
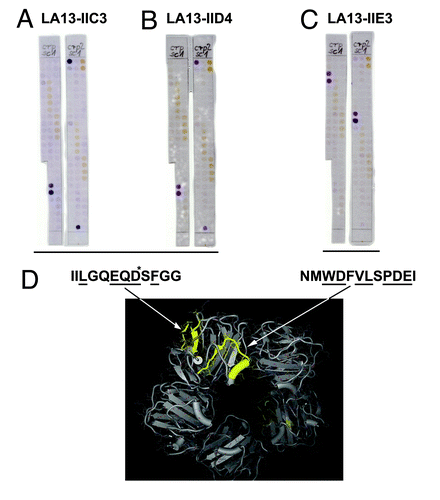
Figure 6. Stability tests of CRP specific scFvs. (A) Long-term stability of the scFv LA13-IIE3 was tested by incubation in PBS for up to 45 d at 37°C. Samples were analyzed by ELISA using CRP as antigen and BSA as control antigen. Freshly thawed scFv samples were used as references corresponding to 100% binding activity. For comparison two scFvs, the lysozyme specific scFv D1.3 and the mucin 1 specific scFv IIB6 were analyzed and tested to their antigens. Bound scFv antibody was detected with mouse α-myc-tag mAb and secondary antibody HRP conjugate. (B) Samples of the CRP specific scFvs LA13-IIE3 and LA13-IID4 were incubated with a final concentration of 3 M GuHCl for 30 min at room temperature followed by dialysis against PBS. The procedure was repeated several times and antigen binding activity was compared with untreated scFv as reference value.
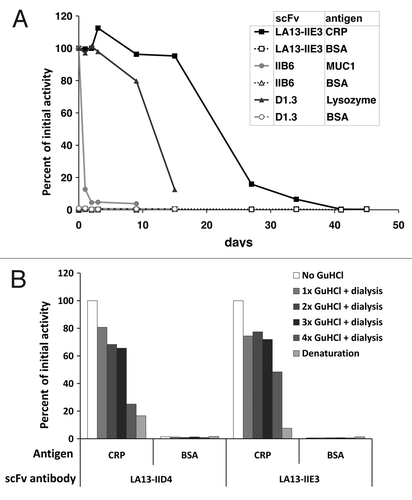
Figure 7. Optimizing conditions for scFv coupling to gold sputtered surface. Conditions for functional coupling of the anti-CRP capture scFv LA13-IIE3 to a gold-sputtered surface of a polypropylene microtiter plate (Gold-PP) were tested by sandwich ELISA using the antigen CRP and for detection anti-CRP mAb followed by a goat-anti mouse-HRP conjugate. Coupling was tested with SAMs using 11-MUA, DSP, or cystamine / glutaraldehyde (Cys/GA), or by direct incubation in combination with (A) different blocking conditions using milk powder (MP), BSA or caseinate or (B, C) by comparing unmodified scFv with the scFv-Cys variant containing a free C-terminal cysteine using (B) caseinate blocking and different SAMs, or (C) Cys/GA coupling and different blocking conditions. (D) Total amount of coupled scFv was detected by myc-tag specific mAb Myc1-9E10 was compared with the amount of functional scFv detected by CRP sandwich ELISA for different SAM coupling (casinate blocking). Polypropylene (PP) microtiter plates were used as control.
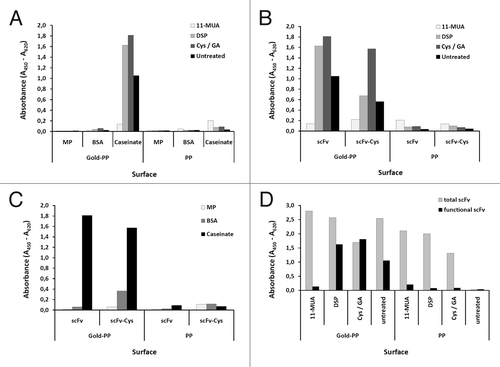
Figure 8. Preparation of the scFv LA13-IIE3 QCM microchip immunosensor and repeated CRP measurements. All injections of scFv, caseinate and CRP are indicated with black arrows whereas those of buffer (PBS) are indicated with gray arrows. The gold sputtered surface of the QCM sensor was activated with a cystamine / glutaraldehyde SAM followed by two injections of 20 and 60 µg/mL scFv LA13-IIE3, respectively. After washing with PBS 10 mg/mL caseinate was injected to block unspecific binding sites. After washing with PBS several measurement cycles were performed comprising injection of 0.25 to 1.0 mg/mL CRP followed by washing with PBS until baseline was retained. A total of 1.0 mg/mL BSA was measured to test unspecific frequency shifts, followed by new CRP measurements. Flow rate was kept at 34 µL/min. Sensor frequency was measured every 83 ms. A total of 20,766,111 Hz was subtracted to set the frequency before scFv coupling to 0 Hz.
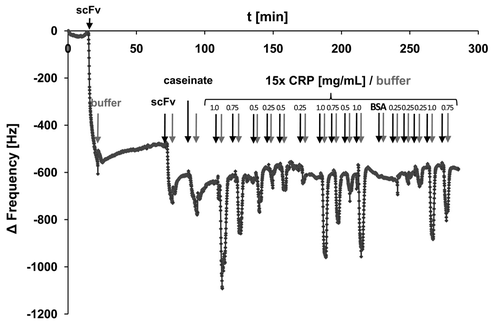
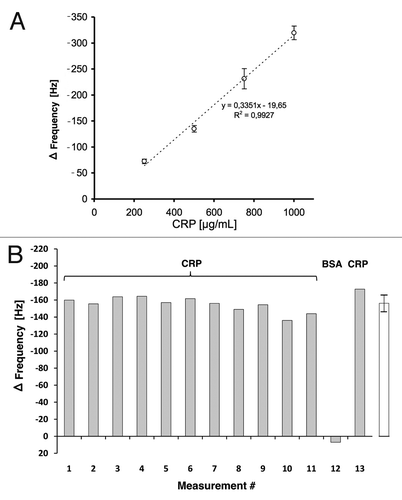
Figure 9. (A) Calibration of the scFv LA13-IIE3 QCM sensor. Measurements disturbed by air bubble peaks were excluded from the calculation of the linear regression equation (dotted line). Correlation coefficient R2 is indicated. Error bars represent standard deviations from triplicate measurements. (B) Repeated measurements to validate reproducibility of CRP specific frequency drops measured with the scFv LA13-IIE3 QCM immunosensor. A total of 12 injections of 0.5 mg/mL CRP and one injection of BSA (#12) used as negative control were applied to the same sensor chip using constant flow rate of 34 µL/min. Sensor frequency was measured every 83 ms. The maximum frequency change before and during CRP or BSA injection is shown. The right (white) bar represents the average frequency shift and standard deviation of all CRP measurements (#1–11 and #13).