Figures & data
Figure 1. Hypothetical successful treatment with omalizumab in a severe asthma cohort. 434 severe asthma patients were stratified for hypothetical eligibility for omalizumab treatments based on atopic status [at least one specific allergen result ≥ 0.35 kU/L from ALT. ALTERNATA, CAT DANDER, COCKROACH,, D. FARINAE, D. PTERONYSSINUS, DOG DANDER and additional panel of allergens dependent on country (Table S1)], IgE/bodyweight eligibility (based on US dosing table; Table S2) and responder status after 16 wk by physician’s global assessment (based on responder rate of 61% in Bousquet et al. 2007Citation12)
![Figure 1. Hypothetical successful treatment with omalizumab in a severe asthma cohort. 434 severe asthma patients were stratified for hypothetical eligibility for omalizumab treatments based on atopic status [at least one specific allergen result ≥ 0.35 kU/L from ALT. ALTERNATA, CAT DANDER, COCKROACH,, D. FARINAE, D. PTERONYSSINUS, DOG DANDER and additional panel of allergens dependent on country (Table S1)], IgE/bodyweight eligibility (based on US dosing table; Table S2) and responder status after 16 wk by physician’s global assessment (based on responder rate of 61% in Bousquet et al. 2007Citation12)](/cms/asset/c45c62e8-00ce-40f6-ba2e-488d05a5e51a/kmab_a_10928394_f0001.gif)
Figure 2. In vitro characterization of MEDI4212. Neutralization of human IgE was assessed in FcεRI-dependent (A, B and C) or CD23-dependent (D and E) assays. (A) Human IgE-FcεRI binding assay; (B) RBL-ER51 calcium signaling; (C) LAD2 β-hexosaminidase release; (D) IM9 binding; (E) U937 phagocytosis of IGROV1 cells. Folate receptor α (FRα). Data points represent average of duplicate determinations from a representative experiment (A) or mean ± SEM of combined data from 3–11 experiments (B–E). All test compounds are IgG unless stated otherwise.
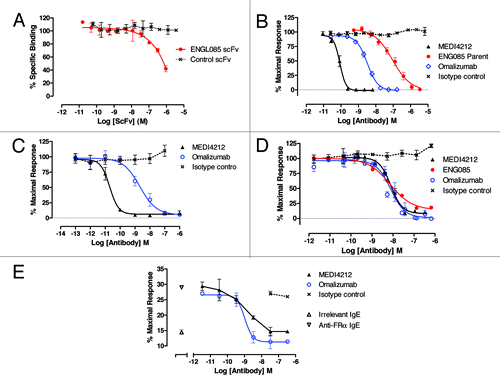
Table 1. Equilibrium dissociation constant (KD) measured by KinExA
Table 2. MEDI4212 Inhibits IgE-mediated functional responses
Figure 3. MEDI4212 does not trigger receptor-bound IgE. Cross-linking of FcεRI-bound IgE by MEDI4212, in comparison with a commercially available polyclonal anti-IgE, was measured by (A) calcium signaling in RBL-ER51 cells (B) human whole blood histamine release assay. (C) Binding to the IgE-loaded CD23 on RPMI-8866 cells was determined for test antibodies in comparison to a commercially available positive control anti-IgE. Data points represent mean ± SEM of combined data from 4–13 experiments (A and B) or average of duplicate determinations from a representative experiment (C).
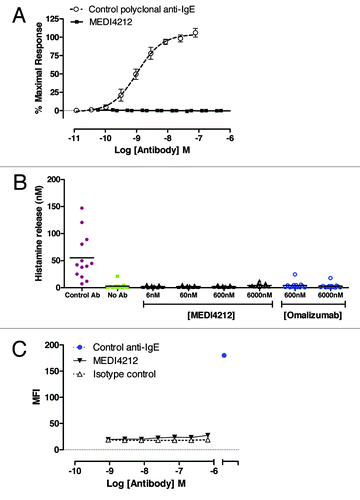
Figure 4. Structural definition of MEDI4212-IgE interaction. (A) Overview of the MEDI4212 Fab and IgE Cε3-Cε4 complex. The two identical IgE chains are colored dark red and salmon and the MEDI4212 fab domains blue, with the heavy chain dark blue and the light chain light blue. (B) A surface representation of MEDI4212 Fab illustrating the large fraction of framework residues in the paratope for IgE. MEDI4212 colored as previously, CDR- not engaged in IgE binding light pink, CDR engaged in binding magenta and framework/Vernier residues engaged in IgE binding in dark blue. (C) Framework residues in MEDI4212 (blue) involved in binding IgE (light red). Arg77 (right panel) forms a salt bridge directly with Glu472 in IgE. Ser75 (left panel) is involved in a stabilizing H-bonding network at the interface to IgE, positioning Asp72 in position to form a salt bridge with Arg342 of IgE. (D) Mode of action of MEDI4212 with respect to FcεRI binding. Overlay of the MEDI4212:IgE complex with the structure of FcεRI:IgE shows a direct overlap of the binding site of MEDI4212 and FcεRI. In addition a large volume, illustrated in orange, describes the steric clash between the two molecules trying to bind simultaneously. (E) Mode of action of MEDI4212 with respect to CD23. Overlay of the CD23:IgE complex structureCitation19 with the MEDI4212:IgE structure shows how the open conformation which IgE adopts in complex with MEDI4212 is incompatible with CD23 binding. The clash volume between CD23 and IgE is illustrated as an orange surface. For clarity, MEDI4212 and IgE of the CD23:IgE complex have been omitted in the picture.
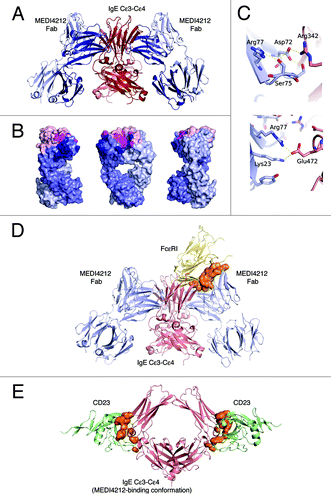
Figure 5. IgE suppression in humans. (A) Suppression of free-IgE in ex vivo human sera from 7 donors with baseline IgE levels 25–1339 IU/mL. Data from each donor at each antibody concentration are plotted as separate data points. (B) Mechanistic PK/PD model. Simulated serum free IgE or specific IgE per basophil profiles following administrations of MEDI4212 and omalizumab in (C) typical asthma patient; (D) High IgE / bodyweight; (E) Low total IgE with high IgE specificity.
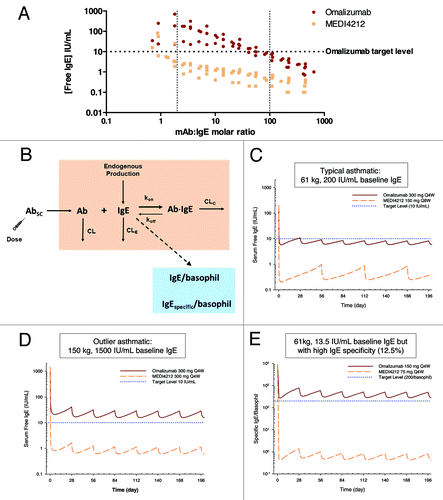
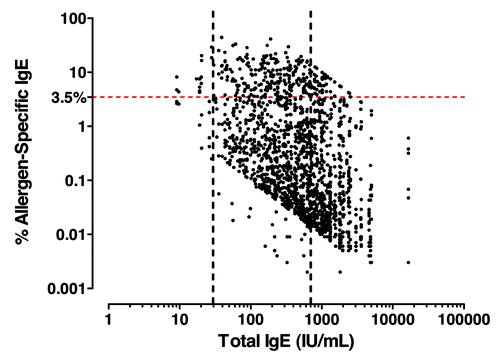
Figure 6. Allergen-specific IgE analysis in severe atopic asthma patients. Total vs. allergen-specific IgE levels in severe atopic asthma. Atopic was defined as least one specific allergen result ≥ 0.35 kU/L from ALT. ALTERNATA, CAT DANDER, COCKROACH, D. FARINAE, D. PTERONYSSINUS, DOG DANDER and additional panel of allergens dependent on country (Table S1).