Figures & data
Figure 1. (A) Molecular formula of the agent (succinylcholine) used as substrate to select the HB2151 E. coli cells able to grow in the selective minimum medium; (B) petri dish containing colonies of HB2151 cells from the first round of selection able to growth in the selective minimum medium.
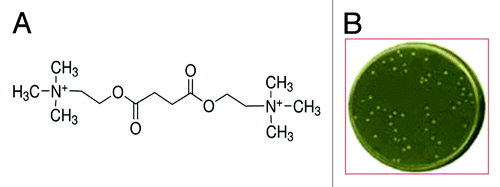
Figure 2. (A) SDS-PAGE of the eluate from HiTrapTM column: a single ~24 kDa protein was detectable; (B) Primary structure of the catalytic scFv as confirmed by MALDI TOF analysis and deposited in GenBank database under the accession number KF914159.
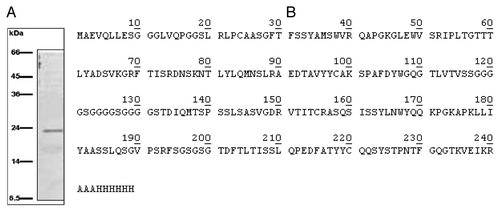
Figure 3. General formula of the thio-substrates, used in the modified Ellman assay (37 °C, pH 7.4) and kinetic curves for the hydrolysis respectively of the three substrates, acetylthiocholine, propionylthiocholine and butyrylthiocholine, by WZ-14.2.1 (10−7 M) in the modified Ellman assay.
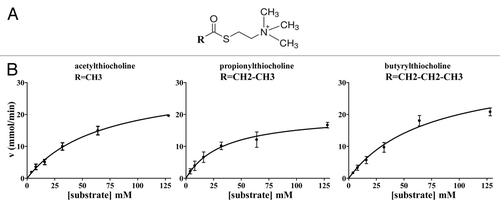
Table 1. Kinetic parameters obtained for the three substrates hydrolysis by WZ-14.2.1 (10−7 M) in the modified Ellman assay (37 °C; pH 7.4)
Figure 5. Concentration-dependent inhibition by the BChE inhibitor ethopropazine on the hydrolysis by WZ-14.2.1 (10−7 M) of the substrate acetylthiocholine verified through the Ellman modified assay (37 °C, pH 7.4). Data are means ± SEM from three separate experiments. *P < 0.05; **P < 0.01
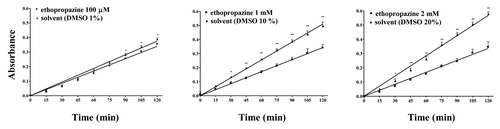
Figure 6. Concentration-dependent inhibition by the Ser blocking agent phenylmethanesuphonyl fluoride (PMSF) on the hydrolysis by WZ-14.2.1 (10−7 M) of the substrate acetylthiocholine verified through the Ellman modified assay (37 °C, pH 7.4). Data are means ± SEM from three separate experiments. *P < 0.05; **P < 0.01
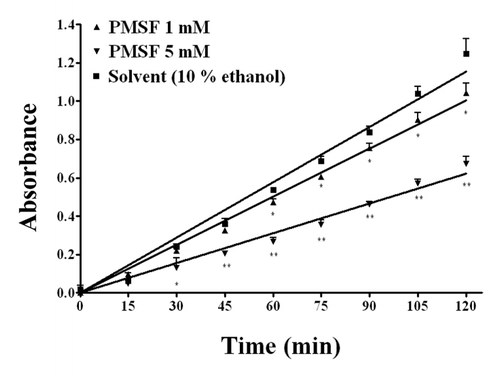
Figure 7. Kinetic profile obtained in the presence and in the absence (control) of WZ-14.2.1 (10−7 M) using the Amplex Red ® fluorimetric acetylcholinesterase/acetylcholine assay; *P < 0.05; **P < 0.01
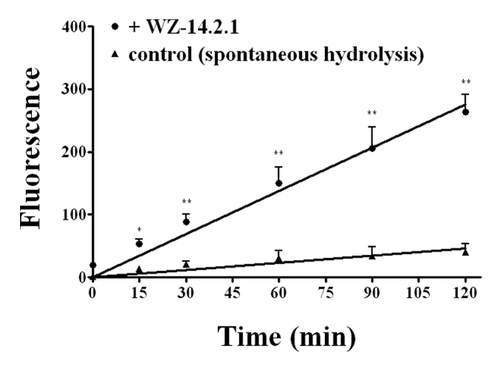
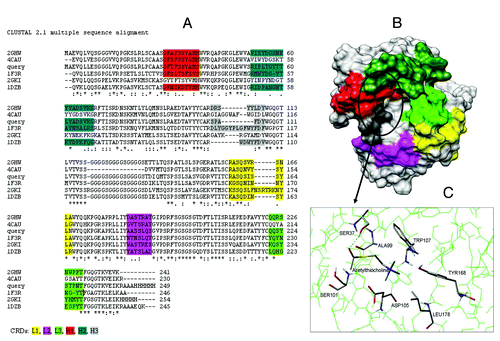
Figure 8. (A) CLUSTALW alignment between WZ-14.2.1 (query) and its closest homologous (identified by BLAST) with the PDB codes 2GHW, 4CAU, 1F3R, 2GKI and 1DZB; (B) 3D model of WZ-14.2.1 obtained by homology building on 2GHW template. The CDR zones (L1, L2, L3, H1, H2, and H3) are colored (in yellow, pink, green, red, dark green, and gray, respectively), the putative binding site is circled; (C) The best conformation of acetylthiocholine in the catalytic antibody obtained with the molecular docking analysis.