Figures & data
Figure 1. Conformation-sensitivity of the epitope recognized by nimotuzumab. Coating extracellular region EGFR recombinant protein was sequentially treated with DTT and iodoacetamide to disrupt disulfide bonds. Binding of nimotuzumab to either treated or non-treated antigen was detected with anti-human IgG antibodies labeled with horseradish peroxidase. Relative antigenicity (%) of denatured erEGFR was calculated taking the reactivity toward the unmodified antigen as the reference. Cetuximab and polyclonal anti-erEGFR monkey antiserum were used as control antibodies.
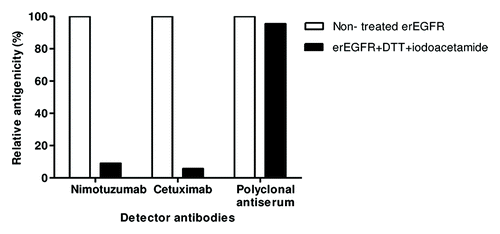
Figure 2. Phage display of EGR domain III and a nimotuzumab-derived antibody fragment. Purified phages displaying either the nimotuzumab target antigenic region (human EGFR Dom III+482–514) or the nimotuzumab binding site itself (a single chain Fv antibody fragment comprising humanized R3 VH and VL regions connected by a linker peptide) were tested by ELISA on microtiter plates coated with different molecules. The anti-tag 9E10 mAb recognizing the c-myc tag fused to all foreign proteins in our system was used to detect the presence of both phage-displayed proteins. Bound phages were detected with an anti-M13 mAb labeled with horseradish peroxidase. A) Phage-displayed EGFR domain III (Dom III+482–514) was tested on nimotuzumab/cetuximab-coated plates. An unrelated antibody was used as negative control. B) Phage-displayed single chain Fv antibody fragment was tested on plates coated with either erEGFR recombinant protein or an unrelated antigen (BSA).
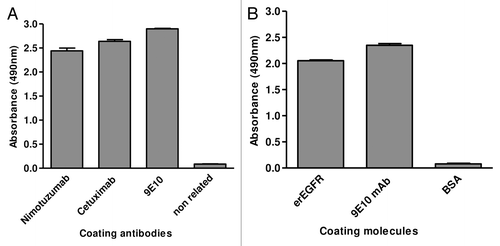
Figure 3. Recognition of phage-displayed EGFR domain III mutated variants. Phages displaying human EGFR Dom III+482–514 mutated variants (where each solvent-exposed residue differing between human and mouse EGFR has been replaced by the aa found in the latter) were produced at a 50 ml scale. Phage-displayed wt Dom III+482–514 was included as a control. Purified phages (1012 viral particles/ml) were incubated on microtiter plates coated with either anti-EGFR mAbs (nimotuzumab [A] and cetuximab [B]) or the anti-c-myc tag 9E10 mAb. Bound phages were detected with an anti-M13 mAb conjugated to horseradish peroxidase. Normalized reactivity for each variant was estimated by dividing the signal obtained with each mAb by the reference signal (measured with the anti-tag mAb). Relative reactivity (%) was calculated as the ratio between normalized reactivity of each variant and that of wt domain III. Arrows indicate lack of recognition of individual variants by a given anti-EGFR antibody.
![Figure 3. Recognition of phage-displayed EGFR domain III mutated variants. Phages displaying human EGFR Dom III+482–514 mutated variants (where each solvent-exposed residue differing between human and mouse EGFR has been replaced by the aa found in the latter) were produced at a 50 ml scale. Phage-displayed wt Dom III+482–514 was included as a control. Purified phages (1012 viral particles/ml) were incubated on microtiter plates coated with either anti-EGFR mAbs (nimotuzumab [A] and cetuximab [B]) or the anti-c-myc tag 9E10 mAb. Bound phages were detected with an anti-M13 mAb conjugated to horseradish peroxidase. Normalized reactivity for each variant was estimated by dividing the signal obtained with each mAb by the reference signal (measured with the anti-tag mAb). Relative reactivity (%) was calculated as the ratio between normalized reactivity of each variant and that of wt domain III. Arrows indicate lack of recognition of individual variants by a given anti-EGFR antibody.](/cms/asset/8f9b840e-e476-4bce-9e24-f056aee362c6/kmab_a_10928915_f0003.gif)
Table 1. Effects of individual replacements within phage-displayed EGFR domain III on recognition by anti-EGFR mAbs
Figure 4. Functional epitope recognized by nimotuzumab. EGFR domain III is shown as a white cartoon with semi-transparent surface. Side chains of the residues contributing to nimotuzumab epitope formation according to mutagenesis studies are represented in red in (A). Relative locations of nimotuzumab and cetuximab epitopes are shown in (B). Domain III residues buried in the interface with cetuximab Fab in the complex (PDB code 1YY9) are colored yellow, while R353, the only residue belonging to both nimotuzumab functional epitope and cetuximab structural epitope, is highlighted in orange. The remaining nimotuzumab epitope residues are colored red. Overlapping between nimotuzumab epitope and ligand binding site is represented in panel (C). The interface between EGFR domain III and EGF and/or TGF-α (residues within 5Å of EGF and TGF-α in receptor/ligand complexes, PDB structures 1IVO and 1MOX respectively) is colored blue. Residues S356, F357, and T358 (highlighted in magenta) are involved in binding of both ligands and nimotuzumab. The rest of the nimotuzumab epitope is shown in red. The figures were generated with Pymol.
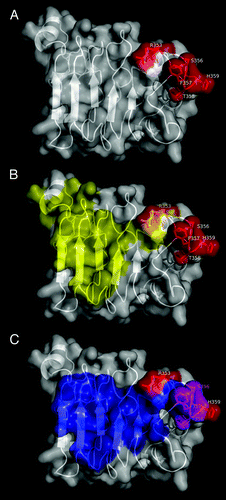
Table 2. Exploration of nimotuzumab paratope through combinatorial and site-directed mutagenesis
Figure 5. Functional map of the nimotuzumab paratope emerging from combinatorial mutagenenesis scanning. The structure of nimotuzumab Fv (PDB code 3GKW) is represented as a cartoon with semi-transparent surface. Heavy chain is shown in white, while light chain is represented in gray. Residues within protruding segments of complementarity determining regions of both chains (which were diversified through combinatorial mutagenesis in phage-displayed libraries) are highlighted in different colors. Targeted VL residues, for which a regular pattern of responsiveness to mutations was not found, are colored yellow. Those VH residues that could be replaced by multiple aa without affecting EGFR recognition are shown in green, while critical functional residues within VH CDR1, CDR2 and CDR3 that were conserved among EGFR-binding positive variants selected on the antigen are colored cyan, purple and dark blue, respectively. The figure was generated with Pymol.
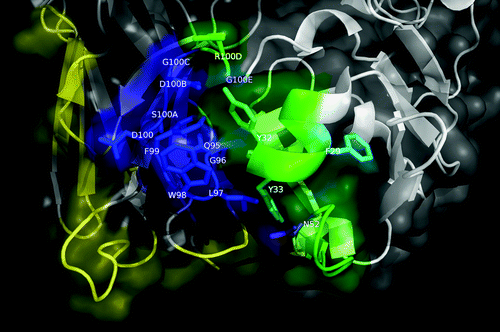
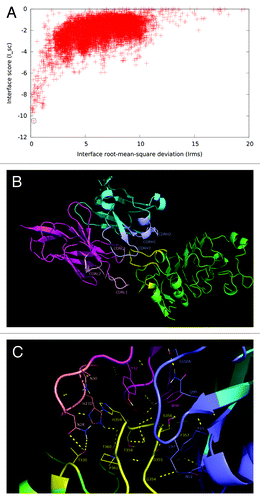
Figure 6. Structural model of nimotuzumab/EGFR complex. (A) Energy landscape of the perturbation analysis from which the best docking model (solution indicated by the dotted line circle) was obtained. (B) Cartoon representation of the structure of the nimotuzumab/EGFR complex according to the selected model. Nimotuzumab heavy and light chains, and erEGFR are colored cyan, magenta and green, respectively. The structural epitope is in yellow while complementarity determining regions from heavy and light chains are represented in blue and red respectively. (C) Interaction interface. Side chains of residues in the interface are shown as lines and hydrogen bonds are shown as discontinuous yellow lines. Residues and chains are colored as in (B).