Figures & data
Figure 1. Non-Viral AAV based single antibody stable pools. (A) Vectors for non-viral AAV expression system. Antibody expression cassettes and a neomycin selection cassette are flanked by AAV ITRs and the p5IEE (pExcel ITRp5). This vector is co-transfected into mammalian cell lines with a plasmid expressing an optimized low level of Rep78 (pRep78low), and stable pools of cells are selected by culture with geneticin. See Methods section for details of vector elements and abbreviations. (B) Optimized AAV-based stable expression is ITR and Rep dependent. Freestyle 293-F cells were transfected with only the antibody expression plasmid with GFP in place of the heavy chain (-Rep) or co-transfected with pRep78low (+Rep). The antibody expression plasmids either contained viral ITR and p5IEE elements (+ ITR and p5IEE; pExcel ITRp5 GFP) or lacked the viral elements (- ITR and p5IEE; pExcel GFP). Data shown are the average percent GFP positive cells as determined by FACS analysis from three time-points of duplicate cultures after transient expression was lost. Error bars represent the standard deviation between two cultures. (C) Single antibody stable pool cultures generated by non-viral AAV based integration express at higher levels than those generated by random integration. The non-viral AAV based pools were generated as described in the Methods section, and the random stable pools were generated identically except that the antibody plasmid did not contain the viral ITRs or p5IEE sequences (pExcel) and was not co-transfected with pRep78low. Frozen vials of AAV-based or random stable pools were thawed and cultured for two weeks. Antibody concentrations from cell culture supernatants were determined by Luminex® immunoassays, and specific productivities (pg/cell/day) were calculated as described in the Methods. Error bars represent the standard deviation between measurements at two time points.
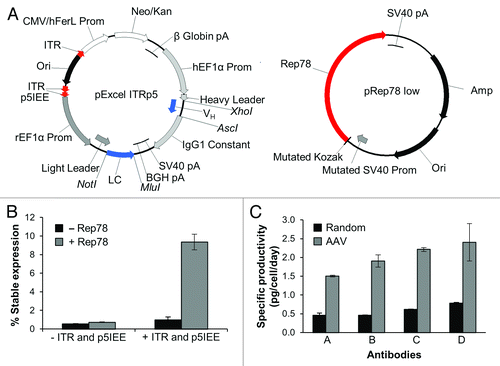
Figure 2. Long-term stability of ACT cultures. Stable pools of cells expressing five different antibodies were mixed and co-cultured. Antibody expression was measured from the cell culture supernatants by Luminex® immunoassays and CIEX chromatography. (A) Fc production is stable for ACT cultures. Data represent ten replicate vials of an ACT composed of five antibodies. Specific productivities (pg/cell/day) were calculated as described in the Methods. Error bars represent the standard deviation from the mean of the 10 cultures. (B) Stability over nine weeks of three replicate vials. Data shown are the percent of each antibody reactivity relative to the sum of all antibody reactivities as measured by Luminex® immunoassays. Error bars represent the standard deviation from the mean of the three cultures. (C) Representative CIEX chromatograms from the beginning, middle, and end of the time course. The chromatogram peaks were assigned by comparison to single antibody expressing cultures analyzed using the same HPLC conditions and method. (D) Stability of 10 replicate cultures over nine weeks. Data shown are the percent of each antibody reactivity relative to the sum of all antibody reactivities as measured by Luminex® immunoassays, where each bar represents a single culture. The error bars represent the standard deviation of Luminex® measurements of the percent of each antibody in the mixture over a nine week period.
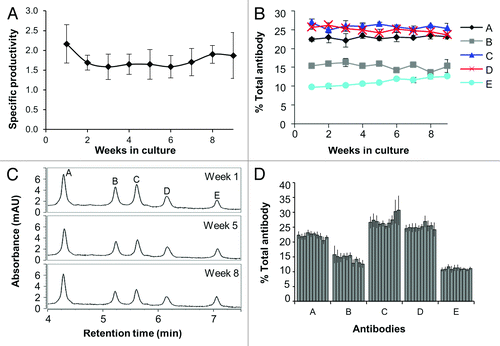
Table 1. Variability between replicate ACT vials and over time in ACT cultures
Figure 3. Stability of complex mixtures composed of 10 antibodies. (A) Sample CIEX chromatograms used for antibody quantitation. The antibody peaks were labeled by comparison with cell culture supernatants from cultures expressing single antibodies using the same HPLC conditions and method. (B) Stability of nine antibodies from the 10 antibody ACT over an eight week period. Data represent percent of each antibody relative to the sum of all antibodies as determined by integration of the area under the curve from the CIEX profiles. Error bars represent the standard deviation from four different thawed vials. (C) The tenth antibody (antibody J) was undetectable by CIEX chromatography, but was measurable by antigen-specific Luminex® immunoassay. The percent of the total Fc in the culture is graphed, and the error bars represent the average of four replicate cultures.
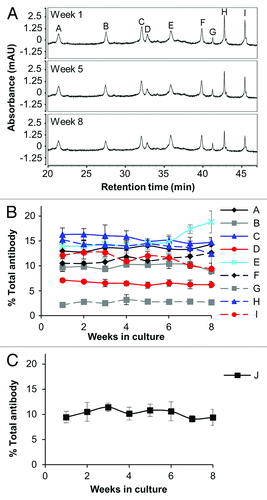
Figure 4. ACT transfection, freezing, and culturing process is reproducible. (A) Diagram of the experiment. Two ACT mixtures were made from independent single antibody stable pools that were generated by independent sets of transfections. Duplicate vials were then thawed, and each vial was split into duplicate cultures. (B) and (C) Long-term stability of two independently made ACTs. The graphs in parts (B) and (C) represent the two independently transfected ACTs described in part A. Relative expression of the three antibodies was measured by Luminex® immunoassays. Error bars represent the standard deviation from the mean of the four replicates of each mixture. (D) Stability of two replicate mixtures, thawed in duplicate and cultured in duplicate over ten weeks. Data shown are the percent of each antibody reactivity relative to the sum of all antibody reactivities as determined by antigen-specific Luminex® immunoassays, where each bar represents a single culture, as diagrammed in part (A). The dark gray bars come from one set of transfections, while the light gray bars come from another. The error bars represent the standard deviation of Luminex® measurements of the percent of each antibody in the mixture over a 10 wk period.
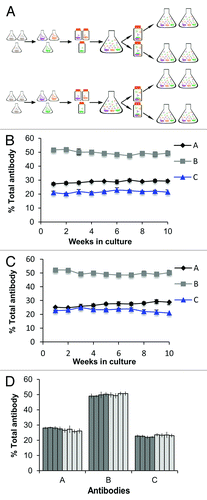
Figure 5. ACT cultures generated by non-viral AAV-based expression are more stable than cultures generated by random integration. (A) AAV generated and (B) Random generated pools produced in parallel. Random cultures were generated by transfection with the antibody expression cassette lacking viral elements (pExcel), while AAV-based cultures were generated by co-transfection with the antibody expression cassette containing the viral ITRs and p5IEE (pExcel ITRp5) and pRep78low. Data shown are the percent of each antibody reactivity relative to the sum of all antibody reactivities as measured by Luminex® immunoassays. Error bars represent the data from four replicate thawed vials for AAV-based cultures (part A) and three replicate thawed vials for Random cultures (part B). (C) Stability of eight replicate ACT cultures over a 12 wk time course. Data shown are the percent of each antibody reactivity relative to the sum of all antibody reactivities as determined by antigen-specific Luminex® immunoassays, where each bar represents a single culture. The error bars represent the standard deviation of Luminex® measurements of the percent of each antibody in the mixture over a 12 wk period. The bars with asterisks were cultured for six weeks.
Figure 6. ACT Expression System Single antibody stable pools are created by co-transfection of pRep78low and pExcel ITRp5 antibody containing plasmids (). These cultures are frozen as single antibody stable pool cell banks, thawed, and productivity for each stable pool is measured. Using the productivity data, single antibody cultures are mixed into one ACT culture containing cells expressing all of the antibodies in the desired combination at the desired concentrations. Master cell banks are made from the ACT cultures and frozen. These cell banks will be transferred to a cGMP facility for the creation of working cell banks for scale up to production bioreactors to create the ACT product.