Figures & data
Figure 1 Fluidic configuration of the 2-dimensional chromatography with online dilution: (A) sample loading; (B) peptide fractionation using the first chromatographic dimension (high pH reversed-phase), and peptide trapping; (C) peptide separation in the second dimension (low pH reversed-phase).
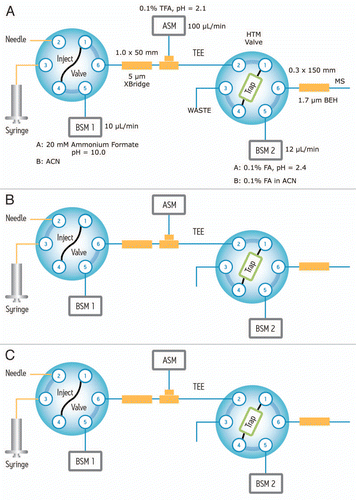
Figure 2 Reproducibility of the 2D-LC setup for four consecutive experiments: (A) extracted mass chromatograms of T49/50 peptide from BSA (DAIPENLPPLTADFAEDKDV(C)K, [MH3]3+ = 820.06) which eluted only in Fraction 3 of 5 step elutions (with 15.4% ACN); (B) extracted mass chromatograms of T43 peptide from ENL (VNQIGTLSESIK, [MH2]2+ = 644.86) eluted only in Fraction 4 out of 5 (using 18.6% ACN). All mass chromatograms were generated using an extraction mass window of 0.1 Da around the corresponding monoisotopic peaks. Second dimension chromatography runs were performed at 12 µL/min using a 30 min gradient (7–35% ACN, 0.1% FA). The amounts of digests loaded on column were 20 fmoles of ENL and 100 fmoles of BSA.
![Figure 2 Reproducibility of the 2D-LC setup for four consecutive experiments: (A) extracted mass chromatograms of T49/50 peptide from BSA (DAIPENLPPLTADFAEDKDV(C)K, [MH3]3+ = 820.06) which eluted only in Fraction 3 of 5 step elutions (with 15.4% ACN); (B) extracted mass chromatograms of T43 peptide from ENL (VNQIGTLSESIK, [MH2]2+ = 644.86) eluted only in Fraction 4 out of 5 (using 18.6% ACN). All mass chromatograms were generated using an extraction mass window of 0.1 Da around the corresponding monoisotopic peaks. Second dimension chromatography runs were performed at 12 µL/min using a 30 min gradient (7–35% ACN, 0.1% FA). The amounts of digests loaded on column were 20 fmoles of ENL and 100 fmoles of BSA.](/cms/asset/5614bf94-0f96-4caa-ae64-b740a47b702e/kmab_a_10918748_f0002.gif)
Figure 3 Chromatographic performance (e.g., RT reproducibility and peak width) is maintained during 1st dimensional fractionation: mass chromatograms of ENL T43 peptide obtained under four fractionation conditions: (A) “simulated” 1D run using a single elution step (from 10.8 to 50% ACN); (B) fraction 2 out of 3 (from 10.8 to 18.6% ACN); (C) fraction 4 out of 5 (from 15.4 to 18.6% ACN); (D) fraction 5 out of 10 (from 15.4 to 16.7% ACN). The data was acquired in continuum mode and all mass chromatograms used an extraction window of 0.1 Da around the corresponding monoisotopic peak. All separations used a 30 min gradient (7–35% ACN, 0.1% FA).
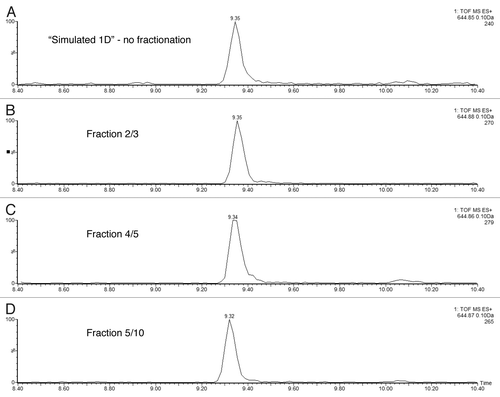
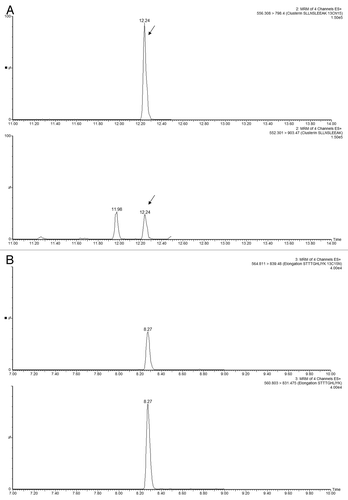
Figure 4 Performance of high-pH peptide fractionation: (A) mass chromatograms of ENL T43 peptide recorded for 3 consecutive fractions during a 10-step 2D fractionation experiment: fraction 4/10 corresponds to a step elution from 14.0 to 15.4% ACN; fraction 5/10 was eluted from 15.4 to 16.7% ACN; fraction 6/10 (16.7 to 18.6% ACN). (B) mass chromatograms of ADH T26 peptide (EALDFFAR, [MH]1+ = 968.48) recorded for 3 consecutive fractions during a 10-step 2D fractionation experiment: fraction 5/10 corresponds to a step elution from 15.4 to 16.7% ACN; fraction 6/10 was eluted from 16.7 to 18.6% ACN; fraction 7/10 (18.6 to 20.4% ACN). The data was acquired in continuum mode and all mass chromatograms used an extraction window of 0.1 Da around the corresponding monoisotopic peaks. All separations were performed using a 30 min gradient (7–35% ACN, 0.1% FA). The amount loaded on-column was 20 fmoles ENL and 2 picomoles ADH digest.
![Figure 4 Performance of high-pH peptide fractionation: (A) mass chromatograms of ENL T43 peptide recorded for 3 consecutive fractions during a 10-step 2D fractionation experiment: fraction 4/10 corresponds to a step elution from 14.0 to 15.4% ACN; fraction 5/10 was eluted from 15.4 to 16.7% ACN; fraction 6/10 (16.7 to 18.6% ACN). (B) mass chromatograms of ADH T26 peptide (EALDFFAR, [MH]1+ = 968.48) recorded for 3 consecutive fractions during a 10-step 2D fractionation experiment: fraction 5/10 corresponds to a step elution from 15.4 to 16.7% ACN; fraction 6/10 was eluted from 16.7 to 18.6% ACN; fraction 7/10 (18.6 to 20.4% ACN). The data was acquired in continuum mode and all mass chromatograms used an extraction window of 0.1 Da around the corresponding monoisotopic peaks. All separations were performed using a 30 min gradient (7–35% ACN, 0.1% FA). The amount loaded on-column was 20 fmoles ENL and 2 picomoles ADH digest.](/cms/asset/4887c329-cd60-42c3-a82e-4db921a6469e/kmab_a_10918748_f0004.gif)
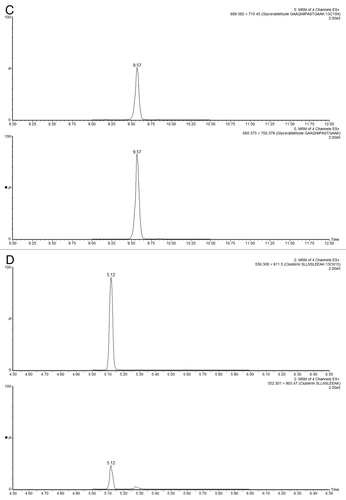
Figure 5 ESI-MS spectra of ENL T43 peptide in a complex peptide background produced by spiking the ENL digest in an E. coli lysate digest. Each spectrum is composed of 10 combined scans across the entire chromatographic peak-width of T43: (A) “simulated” 1D run using a single elution step (from 10.8 to 50% ACN); (B) fraction 2/3 (from 10.8 to 18.6% ACN); (C) fraction 4/5 (from 15.4 to 18.6% ACN); (D) fraction 6/10 (from 16.7 to 18.6% ACN); (E) fraction 6/10 from the ENL digest (no E. coli digest, same 2D fractionation protocol). All separations employed a 30 min gradient (7–35% ACN, 0.1% FA). The amount of ENL digest loaded on column was 20 fmoles for all experiments.
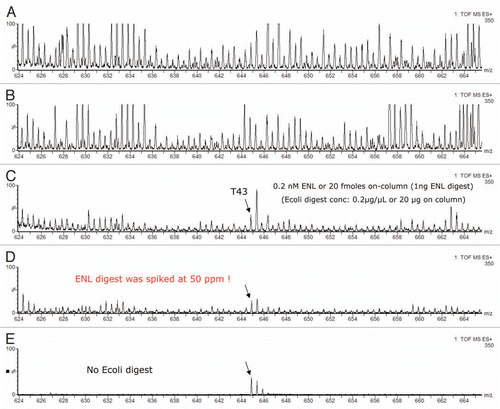
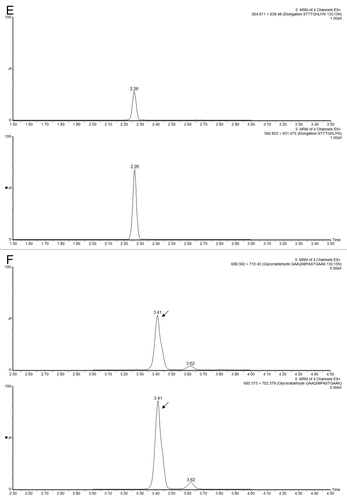
Figure 6 Example of MRM interference: (A) transition 644.9→947.5 of ENL T43 peptide provides a _clean_channel for quantification of this peptide in the PTG1 digest; (B) another transition from the same peptide is obscured by a very strong interference from the sample matrix.
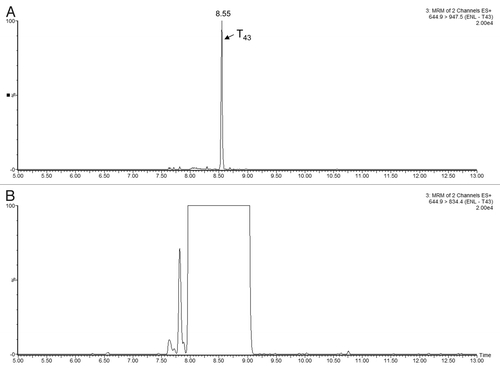
Figure 8 Comparison of HCP quantification between MSE and MRM methods: TOF-based quantification (MSE)is based on the Hi3 method and MRM quantification is based on the peak area from the signal of spiked 13C15N-isotopically labeled peptides with a known concentration. Protein concentrations (ppm) measured in six mAb preparations are shown for (A) clusterin, (B) elongation factor 1α and (C) glyceraldehyde-3-phosphate dehydrogenase.
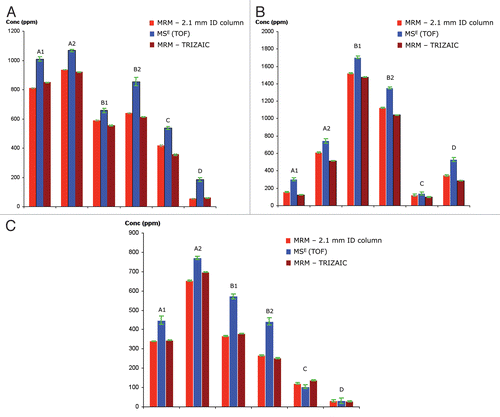
Table 1 HCPs identified across six PTG1 preparations.