Figures & data
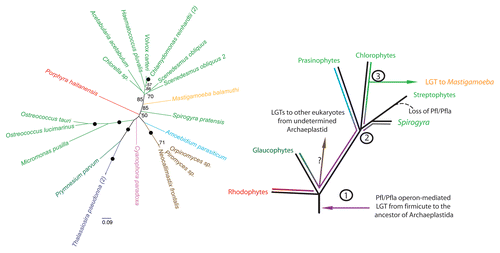
Figure 1 Maximum likelihood (ML) analysis of eukaryotic pyruvate formate lyase (on the left) reveals unique inter-kingdom relationships. RAxML version 7.2.6,28 was used to construct all phylogenetic analyses under the Le and Gascuel (LG)29 amino acid substitution model plus gamma model of rates across sites (denoted PROTGAMMALGF in RAxML). Bootstrap support for bipartitions was estimated from 100 bootstrap replicates and mapped onto the best scoring ML tree (obtained from 20 heuristic search replicates). Branches with 100% bootstrap support are denoted with a black circle, unlabelled branches have support values less than 50%. Organism classifications are indicated in colour: viridiplantae (light green), diatom (purple), amoebozoan (orange), fungi (brown), rhodophyte (red), glaucophyte (pink), icthyosporean (blue) and haptophyte (dark green). The bracketed number represents the number of taxa not displayed. (On the right) Hypothesis for the origin of eukaryotic PFL/PFLAE and its transfer amongst eukaryotes. 1) Operon-mediated transfer of PFL/PFLAE from a firmicute-like bacteria in the common ancestor of Archaeplastida. and subsequent evolution of the eukaryotic PFL over speciation events (purple line). 2) Divergence of the main lineages of green algae and higher plants. 3) Further evolution of the green lineages gave rise to distinct types of PFL/PFLA in the prasinophytes (blue), chlorophytes (green) and early-branching streptophytes (grey). We propose that Mastigamoeba received PFL/PFLAE from a chlorophyte alga while chytrid fungi and other eukaryotes likely acquired the enzymes from a currently unidentified extant or ancestral Archaeplastids.