Figures & data
Figure 1. LPS stimulates tumor-cell adhesion in the liver by disrupting endothelial integrity. (A) Tumor cells in the livers of rats that had been treated with PBS or LPS and were sacrificed 1.5 h after tumor-cell inoculation. Red: DiI-labeled CC531s, blue: cell nuclei, arrowheads point to CC531s cells. (B) Quantification of tumor cells in the livers of rats after PBS or LPS treatment. n = 4 per group; *p < 0.05. (C) ZO-1 staining in the livers of rats that had been treated with PBS or LPS and were sacrificed after 1.5 h. Green: ZO-1; blue: nuclei.
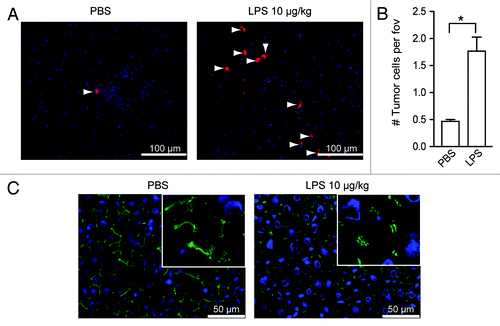
Figure 2. LPS leads to endothelial damage and causes enhanced tumor-cell adhesion. (A) Schematic overview of the co-culture of HUVEC and macrophages (MΦ) in vitro. (B) HUVEC-macrophages co-cultures were incubated with different concentrations of LPS. Red: HUVECs fluorescently stained with Ulex-Rhodamin, blue: cell nuclei. (C) Quantification of damaged endothelial area (%). *p < 0.05; **p < 0.01; ***p < 0.001. (D) Tumor-cell adherence after exposure of HUVEC-MΦ co-cultures to control or LPS. Red: HUVECs, blue: cell nuclei, light blue: SW620 tumor cells. Dotted lines indicate damaged areas without endothelial cells.
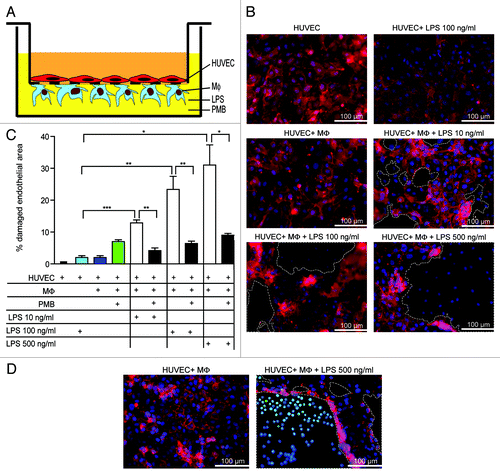
Figure 3. LPS stimulates rapid macrophage activation. (A) Human macrophages were incubated with either PBS or LPS and cells were analyzed by SEM at different time points. (B) Macrophages were treated with PBS or LPS for 2 h and then analyzed by TEM. Representative pictures are shown.
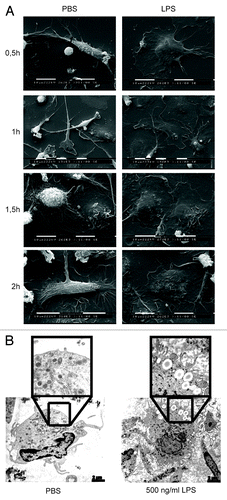
Figure 4. LPS-induced tumor-cell adhesion is Kupffer cell (KC)-dependent. (A) Tumor cells in the livers of control or KC-depleted rats that were treated with LPS. Red: DiI-labeled CC531s, blue: cell nuclei, arrow heads point to CC531s cells. (B) Quantification of tumor cells in the livers of control or KC-depleted rats that were treated with LPS. n = 4 per group; ***p < 0.001 (C) ZO-1 staining in the livers of control or KC-depleted rats that were treated with LPS. Green: ZO-1, blue: cell nuclei.
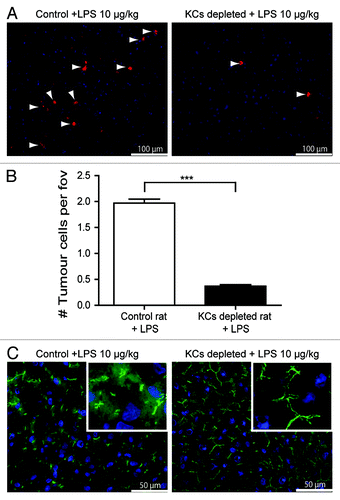
Figure 5. LPS-induced damage is mediated by ROS. (A) Co-culture of HUVECs and macrophages (MΦ) were incubated with 100 ng/mL LPS in the presence or absence of catalase and/or SOD. Red: HUVECs, blue: cell nuclei. Dotted lines indicate areas without endothelial cells. (B) Quantification of damaged endothelial area (%). *p < 0.05; **p < 0.01.
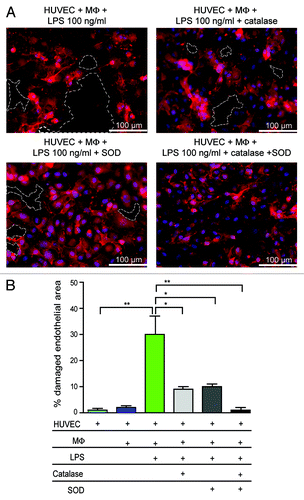
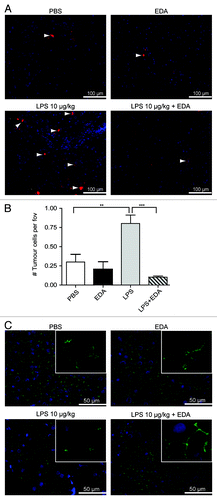
Figure 6. LPS-induced tumor-cell adhesion in the liver is prevented by treatment with edaravone (EDA). (A) Tumor cells in the livers of rats that were treated with PBS, EDA, LPS or LPS plus EDA. Rats were sacrificed 1.5 h after tumor-cell inoculation. Red: DiI-labeled CC531s cells, blue: cell nuclei, arrow heads point to CC531s cells (B) Quantification of the tumor cells in the livers of rats that were treated with PBS, EDA, LPS or LPS plus EDA. n = 4 per group; **p < 0.01; ***p < 0.001. (C) ZO-1 staining in the livers of rats that were either treated with PBS, EDA, LPS or LPS plus EDA. Green: ZO-1, blue: cell nuclei.