Figures & data
Figure 1. Flow cytometry-based detection of epitope-bound monoclonal TCRs on peptide-pulsed T2 cells. T2 cells were pulsed with the indicated HLA-A2-restricted peptides in concentrations ranging from 10−5 to 10−9 M. Epitope presentation was detected using a high-affinity biotinylated monoclonal T-cell receptor (mTCR) specific for gp100 (left panel) or Wilms’ tumor 1 (WT1, right panel) and flow cytometry, upon staining with phycoerythrin (PE)-conjugated streptavidin. A control measurement was made using T2 cell pulsed with 10−5 M of an irrelevant peptide (shaded gray).
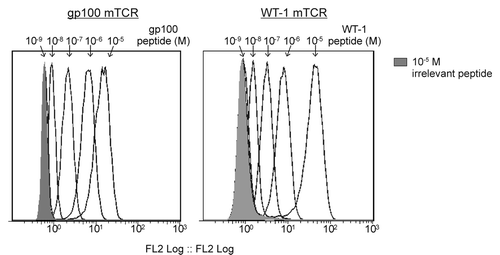
Figure 2. Quantification of epitopes on T2 cells using monoclonal TCRs and single-molecule microscopy. Peptides derived from the tumor-associated antigens (TAAs) gp100, telomerase reverse transcriptase (TERT), and Wilms’ tumor 1 (WT1) were used to pulse T2 cells in concentrations ranging from 10−7 to 10−10 M. Epitopes were bound by high affinity, biotinylated monoclonal T-cell receptor (mTCRs) and visualized by microscopy upon staining with phycoerythrin (PE)-conjugated streptavidin. Solid bars depict the number of epitopes presented by individual T2 cells (means). At concentrations of 10−7 M, the number of epitopes was incompatible with accurate counting. In each case a background measurement was made using a high-affinity mTCR specific for an irrelevant, HIV-1-derived antigen (Gag77–85), using a peptide concentration of 10−5M. The Y-axis on each graph has been optimized to best represent the distribution. Representative phase contrast and fluorescence images are shown for gp100 (upper panel), TERT (middle panel), and WT1 (lower panel). Fluorescence images are three-dimensional reconstructions of individual planes. The brightness/contrast of individual phase contrast and fluorescence images was adjusted to optimize epitope visualization. Scale bar = 20 µm.
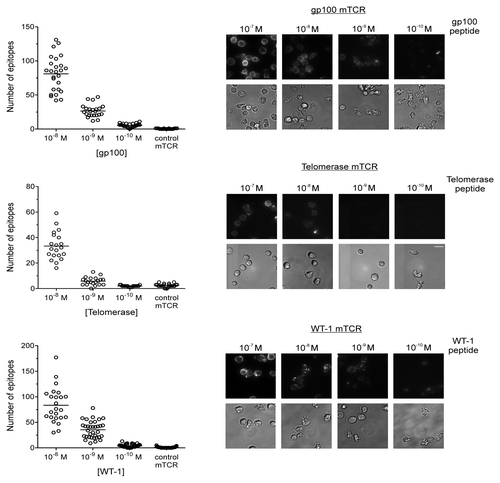
Figure 3. Quantification of epitopes on Mel526 and Mel624 melanoma cells. A gp100-specific biotinylated monoclonal T-cell receptor (mTCR) was allowed to bind to gp100-derived epitopes naturally presented on melanoma Mel526 and Mel624 cells, and visualized by microscopy upon staining with phycoerythrin (PE)-conjugated streptavidin. Solid bars depict the number of epitopes presented by individual tumor cells (means). A background control measurement was made using a high-affinity mTCR specific for an irrelevant, HIV-1-derived antigen (Gag77–85). Representative phase contrast and fluorescence images for the 2 cell lines are shown. Fluorescence images are three-dimensional reconstructions of individual planes. The brightness/contrast of fluorescence images was adjusted to optimize epitope visualization. Scale bar = 20 µm.
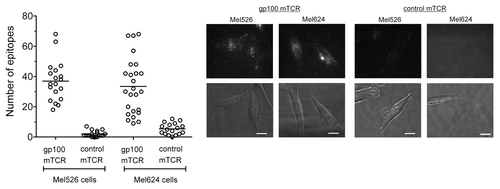
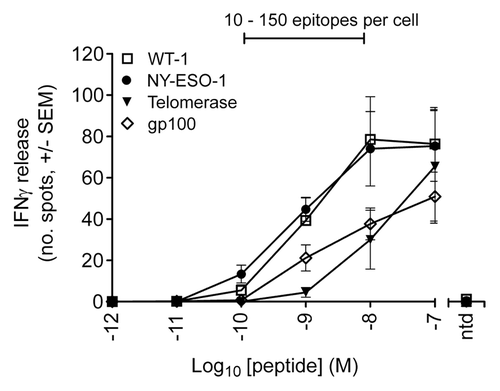
Figure 4. T-cell response to peptide pulsed T2 cells as determined by interferon γ release. ELIPOST assays were used to measure the level of T-cell activation, assessed in terms of interferon γ (IFNγ) release, in response to T2 cells presenting varying levels of epitopes. CD8+ and CD4+ T cells were isolated from peripheral blood mononuclear cells and transduced with wild-type TCRs specific for NY-ESO-1-, gp100-, telomerase reverse transcriptase (TERT)-, and Wilms’ tumor 1 (WT1)-derived peptides. T2 cells were pulsed with antigenic peptides in concentrations ranging from 10−7 to 10−12 M. A control was performed in each case using non-transduced (ntd) T cells and a peptide concentration of 10−7 M. The bar above the graph indicates the peptide concentration range in which physiological numbers of epitopes are presented.