Figures & data
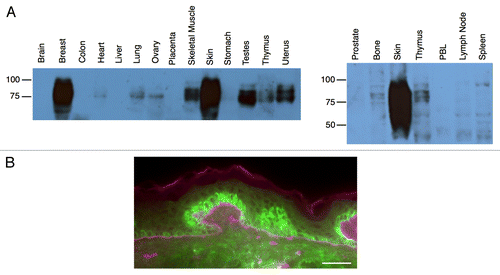
Figure 1. A schematic view of the human papillomavirus life cycle. Daughter cells of epithelial stem cells divide along the basement membrane and then mature vertically through the epithelium without further division (right side, normal epithelium). Stem cells in the basal layer of the epithelium become exposed to human papilloma virus (HPV) as a result of microabrasion or foreign body introduction (left side, infected epithelium). Expression of HPV genes is tightly regulated and linked to epithelial differentiation. Upon infection, viral genomes are established in the nucleus as episomes. Early viral proteins deregulate cell cycle control, allowing viral genome amplification in cells that normally would have exited the cell cycle. The late phase proteins encapsidate newly synthesized viral genomes and mature virions are shed from the most superficial layers of the epithelium. Adapted from Frazer, 2004.Citation109
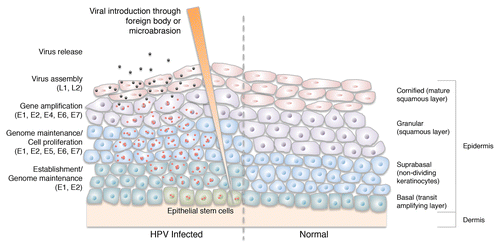
Figure 2. UV radiation (UVR) mutates keratinocyte DNA and inhibits apoptosis of UV-damaged cells through downregulation of FasL and TRAIL. It also induces immunosuppressive IL-10 expression, which inhibits dendritic cells in the skin, suppressing the skin’s immune system. UV-induced proinflammatory cytokines and p53 expression modulate the promoter activity of a number of HPV types. HPV further prevents DNA damage repair by promoting degradation of p300, inhibition of XRCC1 and a reduction in ATR. HPVs also limit apoptosis through proteolytic degradation of pro-apoptotic Bak and upregulation of osteoprotegerin and IL-6. Over time, these interactions result in the accumulation and propagation of somatic mutations, and ultimately, tumorigenesis. FasL: Fas ligand/CD95 ligand, TRAIL: tumor necrosis factor (TNF)-related apoptosis inducing ligand, IL-10: interleukin 10, XRCC1: X-ray repair cross-complementing protein 1, ATR: ataxia telangiectasia-mutated (ATM) and Rad3-related kinase.
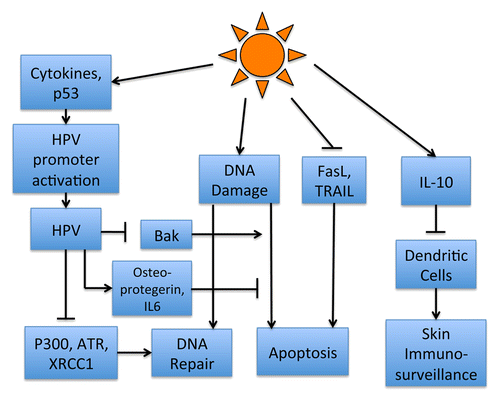
Figure 3. Unbiased protein homology modeling of TMC8 reveals structural similarities with subunit 1 of Cytochrome Ba3 Oxidase. Crystal structure representation of Cytochrome BA3 Oxidase (2QPE) from Thermus thermophiles (left); 2QPE Chain A (blue), 2QPE Chain B (red), and 2QPE Chain C (green). TMC8 reverse homology modeled using CPHmodels-3.2Citation93 (middle, yellow), and overlaid with 2PQE Chain A (right).
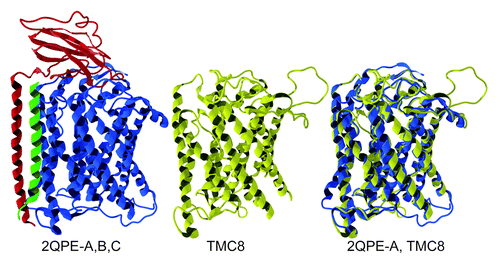
Figure 4. TMC8 protein expression. Expression analysis of transmembrane channel-like 8 (TMC8) by western blot (A) and immunofluorescence staining (B). (A) Human tissue lysates (15 μg, BioChain) were boiled in reducing sample buffer, resolved by 10% SDS-PAGE, transferred to PVDF membrane and probed with an anti-TMC8 antibody (Bethyl Labs). (B) Frozen human skin section (Biochain, CA) stained with antibodies against TMC8 (Bethyl Labs, green) and basal lamina marker laminin (US Biological, purple). Scale bar: 200 µm.