Figures & data
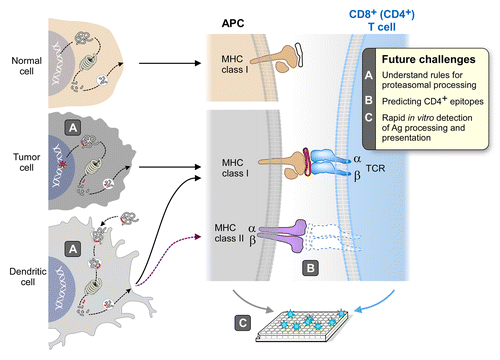
Figure 1. The neoantigen vaccine immunotherapeutic concept. Cancer cell genomes are now known to contain many mutations, some of which create amino acid changes in the encoded proteins. Some of these modified proteins will be partially degraded by the normal cellular re-cycling machinery, creating short 8 to 12 amino acid peptides, and some of these peptides will bind to one of the class I MHC molecules of the individual. Peptide binding to MHC is a critical gateway to both the initiation of a T-cell immune response by the antigen presenting cell (APC), and to the detection and elimination of tumor cells presenting the particular peptide by the stimulated cytotoxic T lymphocyte (CTL). The attraction of neoantigens as cancer targets for the immune system results from the structural and geographical features of the mutation. Central tolerance is known to purge the vast developing T-cell population in the thymus of high avidity T-cell receptors (TCRs) that recognize MHC complexes with native peptides (‘self’). In contrast, high avidity T cells are not centrally deleted against mutated peptide/MHC complexes (like viral antigens) because the mutated antigens are not present in thymus during T-cell development. These high avidity cytotoxic T cells can be selectively amplified and stimulated to attack and kill cells that present mutated peptides. Since neoantigen peptides are only found in tumor cells, the CTLs should show exquisite specificity, reducing the opportunity for autoimmune disease. Desired improvements in the process are shown in the box in the upper right.