Figures & data
Figure 1 Hox genes set and maintain body plan and identity during development. Embryogenesis initiates with the developmental cues provided by maternal gene products that set off zygotic transcription in predetermined patterns. Coordinated expression of the Hox genes (depicted as colored patterns defining expression domains) establishes positional and segmental identity and is maintained throughout organogenesis. The domains of Hox expression profiles are tightly regulated by the PcG and trxG complexes and define the developmental plan of the adult (adapted from ref. Citation130).
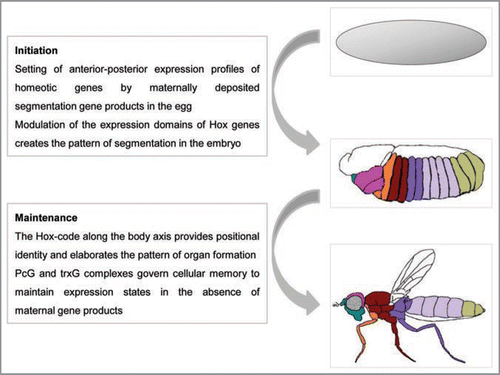
Figure 2 Schematic representation of the action of PcG and trxG complexes on chromatin. Multiple DNA binding proteins (colored circles) interact cooperatively with each other and with the PREs (brown box) located within or near the promoters of Polycomb target genes, setting the appropriate context for assembly of polycomb (PRC1/PRC2) or trithorax (TRX) remodeling activities. Balanced presence of both activating and repressive complexes can create a ‘poised’ state in some genes (dashed arrow) till appropriate differentiation signals are received. Depending on the type of developmental cues, the transcriptional status of the gene is resolved differently. PRC2 causes trimethylation of lysine 27 of histone H3 (H3K27me3; inverted red triangles) leading to recruitment of PRC1 and other modification factors that create a repressed chromatin conformation to keep the gene inactive. Binding of trithorax complexes causes trimethylation of lysine 4 (H3K4me3; inverted green triangles) and recruitment of transcription factors and activators that create a cascade of activating modifications leading to gene transcription.
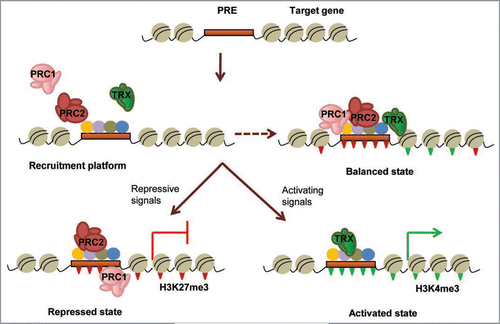
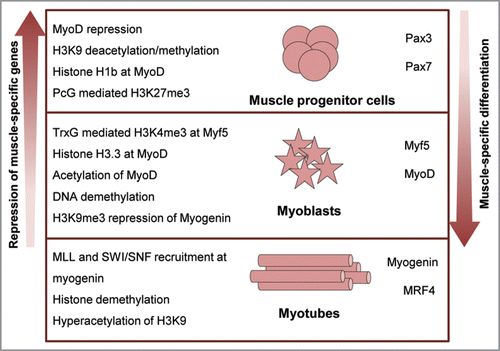
Figure 3 Epigenetic regulation of myogenic differentiation. Muscle myofibres are formed by the activation of the myogenic program in undifferentiated progenitor cells. Terminal differentiation into myotubes involves a series of chromatin remodeling and histone modifications (left) that lead to the derepression of myogenic regulators and the progressive activation of muscle-specific gene cascades (right).