Figures & data
Figure 1. RNA helicases in the spliceosome assembly/disassembly pathway. The order of assembly of U snRNPs and the steps of association and activity of helicases are shown on a transcript containing a single intron. Names of complexes refer to the human nomenclature.
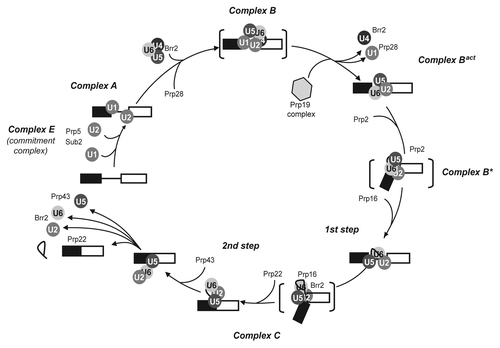
Figure 2. Splicing helicases belong to three distinct families. Primary sequence alignment of S. cerevisiae helicases from the DEAD-box (A), DEAH-box (B) and Ski2-like (C) families involved in pre-mRNA splicing. Black and gray blocks represent the conserved regions within each family. The lines in the amino-termini of DEAH-box helicases indicate the lack of conserved sequences in this region. The positions of the conserved motifs are indicated by vertical rectangles. For DEAH-box and Ski2-like helicases, a downward triangle indicates the position of the β-hairpin proposed to act as a strand separator. Dashed boxes indicate the conserved domains in the carboxy-termini of DEAH-box proteins and in the two helicase modules of the Ski2-like Brr2 helicase. For clarity of the figure we refer to the old nomenclature for conserved motifsCitation19 although a new nomenclature has been proposed.Citation17
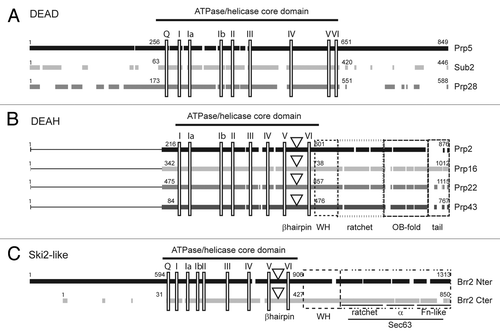
Figure 3. Cytoscape (http://www.cytoscape.org) representation of the interactome of S. cerevisiae splicing helicases. Splicing interactomes of (A) Sub2; (B) Prp5; (C) Prp28; (D) Brr2; (E) Prp2; (F) Prp16; (G) Prp22; (H) Prp43. The list of interactors was obtained from biogrid (http://thebiogrid.org). Only splicing factors are shown, grouped as in ref. Citation1. Colored shapes indicate sub-complex associations of splicing factors. Connectors are colored according to the experimental system used: blue lines represent affinity-capture followed by identification of the prey by mass spectrometry or western blotting, dashed purple lines represent co-fractionation or co-purification experiments, dashed green lines represent genetic interactions and sinusoidal red lines represent yeast two-hybrid interactions.
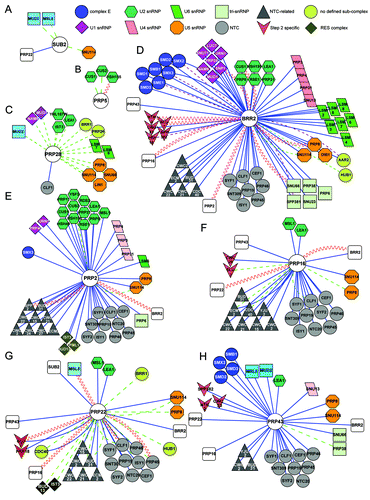
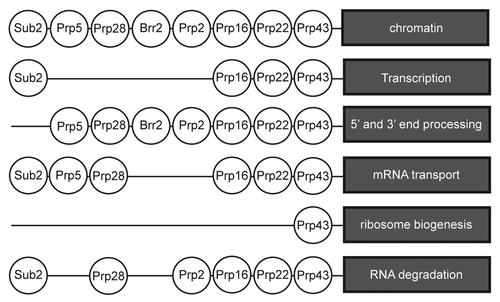
Figure 4. Splicing helicases are connected with other pathways of RNP metabolism. Based on large scale physical and genetic screens (see text), most splicing helicases are connected with several RNP biogenesis events. Only in few cases (see text) has the biological relevance of the proteome data been validated.