Figures & data
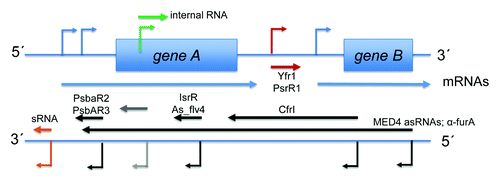
Figure 1. Global analyses for the identification and characterization of sRNAs in R. sphaeroides. (A) dRNA-seq and RNA-seq from Hfq coIP experiments identified intergenic and antisense transcripts representing putative sRNAs and asRNAs, respectively.Citation24,Citation82 Newly identified transcripts and ORF annotations were used for microarray design. The sRNAs given below the colored boxes indicate their identification in the respective analysis and were chosen due to their oxygen-responsive expression (see main text and for details). The heat map illustrates results for these sRNAs from a microarray study conducted for the singlet oxygen (1O2) stress response.Citation21 Expression levels after treatment with methylene blue in light are compared with expression levels before singlet oxygen stress. (B) Integrated Genome Browser (IGB) screenshots for RNA-seq results from Hfq coIP experiments. Mapping of cDNA reads is shown for the coIP with 3xFLAG-tagged Hfq (Hfq coIP) and an unspecific control experiment (control). The scale on the left hand side of each screenshot displays relative read numbers. Red and gray arrows represent sRNA genes and ORFs, respectively. The numbers at the top indicate enrichment factors calculated for sRNAs. Transcript lengths are given for each sRNA.
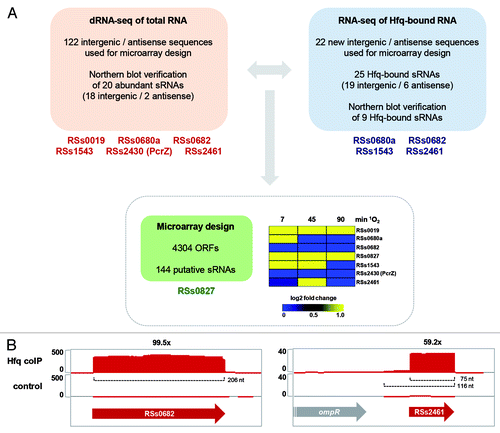
Figure 2. Regulation model for oxygen responsive sRNAs in R. sphaeroides. Reactive oxygen species are either generated by energy transfer during photosynthesis or due to electron transfer mediated by flavoenzymes during aerobic respiration. Several alternative sigma factors (RpoE, RpoHI, RpoHII) are activated, which subsequently induce sRNAs. During aerobic respiration, phosphorylation of the response regulator PrrA by the sensor kinase PrrB is inhibited due to electron flow through the ccb3 oxidase. Under low oxygen tension, PrrA is phosphorylated and triggers PcrZ transcription, which in turn, represses photosynthesis gene expression. Interaction of some sRNAs with Hfq is indicated. (LH, light harvesting complex; RC, reaction center; UQ, ubiquinone pool; Cyt, cytochrome; P, phosphate, DH, dehydrogenase; FADH2, reduced flavin adenine dinucleotide; FFL, feed-forward loop; 1O2, singlet oxygen; O2‒, superoxide radical; H2O2, hydrogen peroxide).
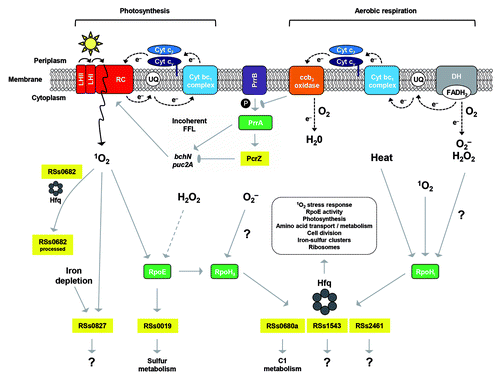
Table 1. Individual sRNA and asRNA regulators discussed in this review
Table 2. Overview on the number and types of putative TSS (see for explanation of TSS types) mapped for photosynthetic model bacteria by dRNA-seq
Figure 3. Summary of demonstrated asRNA and sRNA functions in cyanobacteria. TSS driving the transcription of annotated genes (gTSS, blue), putative sRNAs (nTSS, red), and asRNAs (aTSS, black), together with intragenic TSS sequences (iTSS, green) are visualized by the bent arrows. Details of the TSS numbers and classification mapped in dRNA-seq studies in different cyanobacteria are summarized in . The sRNAs with an at least partially characterized function (symbolized by straight arrows in dark red) are PsrR1 and Yfr1. These are specific trans-acting regulators of gene expression but there are also hundreds of additional sRNAs awaiting more detailed characterization (light red arrows). Relatively short intragenically located asRNAs can act as repressors of gene expression, illustrated by IsrR and As1_flv4 in Synechocystis 6803 (black arrows).Citation61,Citation65 Another type of antisense repressor is represented by α-furA, which in Anabaena 7120 controls the expression of the furA gene. This 2.2 kb-long asRNA originates by read-through from the downstream located gene alr1690.Citation76-Citation78 Longer and shorter asRNAs may also support a high level of gene expression as speculated for the asRNA CfrI of cyanophage S-PM2,Citation72 infecting marine Synechococcus, demonstrated for PsbA2R and PsbA3R in Synechocystis 6803Citation66 or, in the form of several kb long asRNAs in Prochlorococcus MED4, may even protect mRNAs from the RNase E activity enhanced upon infection by the cyanophage P-SSP7.Citation75 Last but not least, there are myriads of intragenic transcripts (green) and asRNAs (gray) whose functions are still entirely unknown.