Figures & data
Figure 1. Complex life cycle of streptomycetes. (A) Morphological diversity of different Streptomyces strains. (B) The life cycle of S. coelicolor. Monogenomic spores of S. coelicolor germinate and grow into the soil. When nutrition becomes limited the vegetative mycelia digest themselves to serve as substrate for aerial mycelia growing out of the soil. Those aerial hyphae differentiate into spores again through insertion of septa.
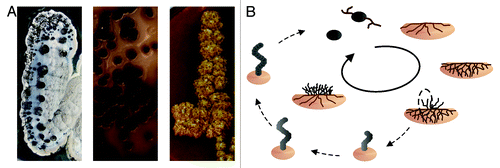
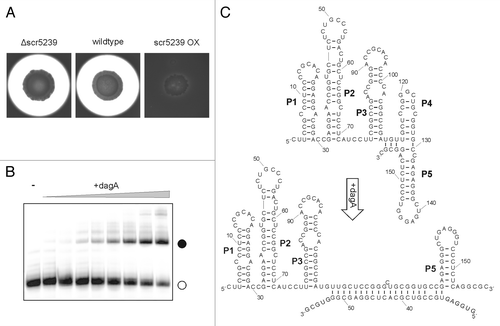
Figure 2. Agarase DagA controlled by scr5239. (A) Agarase assay of S. coelicolor wild-type, scr5239 deletion, and overexpression strain. Agarase expression is visualized as a pale halo around the colony. (B) Electrophoretic mobility shift of radiolabelled scr5239 (open circle) incubated with an increasing amount of a 100 nt mRNA fragment containing the target site of dagA. Complex formation (closed circle) was visualized by gelelectrophoresis. (C) Secondary structure of scr5239 alone (top) and in complex with the mRNA target site (bottom). The structure was calculated using RNAfoldCitation29 and confirmed by structural probing. Stems (pedesand = P) are numbered, the 5′ and 3′ end of each RNA is indicated.