Figures & data
Table 1. Annotated small heat shock genes in selected cyanobacteria from various habitats
Figure 1. Transcriptional analysis of avashort und avalong genes from A. variabilis. (A) Northern analysis of RNA isolated from A. variabilis kept at 23 °C or heat shocked to 40 °C for 1 h. Avashort and avalong mRNA was detected with specific digoxygenine-labeled RNA probes. (B) Sequence alignment of avashort/alrshort and avalong/alrlong. Sequence alignment of the small heat shock gene promoter regions and 5′UTRs from A. variabilis ATCC 29413 (avashort, avalong) and the homologous genes (alrshort, alrlong) from Nostoc PCC 7120 performed using ClustalW2.Citation84,Citation85 Similarity is indicated by asterisks. For avashort and avalong, promoter sequences were computed with BPROM from the softberry webserver, transcriptional start sites (*) were determined by 5′RACE. For Nostoc, start sites were published before.Citation3 Both sequence alignments reveal high similarity. Promoter sequences (-35, -10), transcriptional start sites (*), Shine-Dalgarno sequence (SD), and ATG start codon are depicted.
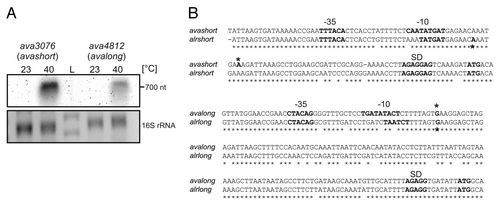
Figure 2. Temperature-dependent translational regulation of reporter activity. (A) Schematic drawing of translational fusion of RNAT of interest to bgaB (thermostable β-galactosidase) in pBAD2-bgaB.Citation29 (B) Reporter gene assay of translational fusions of cyanobacterial RNAT candidates to bgaB tested in E. coli at 30 °C (white columns) or 42 °C (black columns). The well-studied hsp17-RNAT from SynechocystisCitation14 served as positive control. Both the fusion from Anabaena (avashort/avalong) as well as those from Nostoc (alrshort/alrlong) exhibited temperature-induced reporter activity after heat shock from 30–42 °C. The presented results are mean values of a double measurement; mean standard deviation is indicated by error bars.
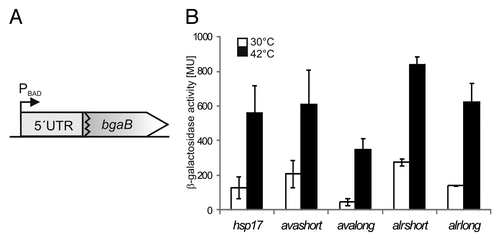
Figure 3. Reporter activity over temperature range. To elucidate the temperature-dependent RNAT activity of the avashort and the avalong-5′UTRs, reporter activity of both RNAT-bgaB fusions as well as the control hsp17-bgaB were analyzed in E. coli over a temperature range from 30–42 °C in 2 degree steps. The presented results are mean values of a double measurement; mean standard deviation is indicated by error bars. For normalization, the respective 30 °C value was set to 1.
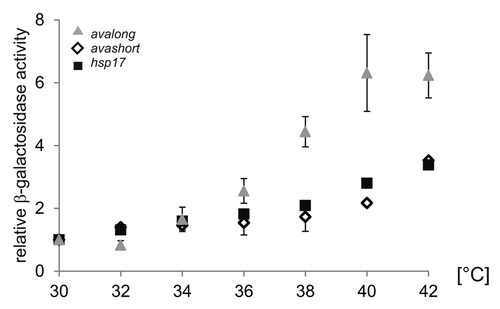
Figure 4. Temperature-dependent structural alterations of avashort-RNAT. (A) Secondary structure prediction of the avashort-5′UTR computed with mfold,Citation86 including 72 Nt coding region (cdr) as well as the first three Nt of the blunt end restriction site EcoRV. SD and AUG are highlighted by gray circles. Black encircled Nt are referred to in the quantification in (C). The 5′ structure forms three hairpins (H1-H3), the SD sequence is partly base paired in H2, and the AUG is embedded in H3. The cdr forms H4 and H5. (B) In vitro transcribed avashort RNA (avashort-5′UTR and 72 Nt) was 5′ labeled and partially digested with RNase T1 (0.001U) and RNase V1 (0.0025U) at the indicated temperatures. For orientation, an alkaline ladder (LOH) was loaded; for the control (C) enzyme was replaced by A. dest. (C) Quantification of altered RNase T1 cleavage intensity between 25–42 °C for selected guanines. Band intensities were quantified from (B) using the Alpha Ease software (Alpha Innotech, Biozym). Shown are relative intensities for each Nt position with band intensity at 30 °C set as 1.
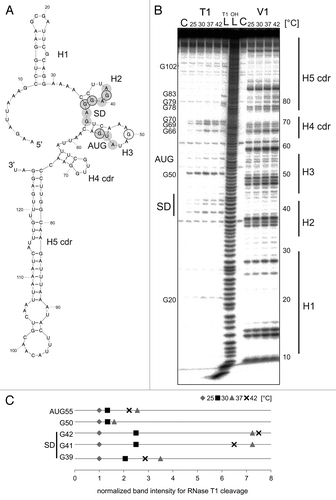
Figure 5. Reporter assay of avashort-bgaB WT and variants. (A) The putative anti-SD forming cytosines C34 and C35 were replaced by UA, UU, GA, and GG, respectively using randomized site-directed mutagenesis. The avashort WT exhibited a 3.5-fold induction of reporter activity after shift from 30–42 °C. All four variants exhibited WT-like activity. (B) Binding of AUG was loosened by changing the G55-C46 pair into a G55-A46 mismatch. Analyzed in the reporter assay, replacement of the cytosine by adenine resulted in complete destabilization of the avashort-RNAT structure visible in almost equal galactosidase activity levels at 30–42 °C. Both the results shown in (A and B) are mean values of a double measurement; mean standard deviation is given by error bars. WT, wild-type.
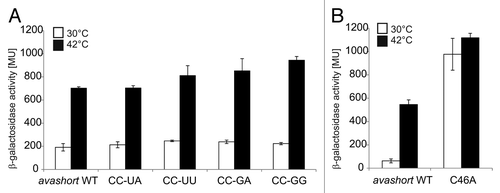
Figure 6. Temperature-dependent structural alterations of avalong-RNAT. (A) Secondary structure prediction of the avalong-5′UTR as well as the first three Nt of the blunt end restriction site EcoRV computed with mfold.Citation86 The structure exhibits two RNA hairpins (H1 and H2), the first possesses a large terminal loop containing a putative alternative anti-SD sequence (marked in gray, asterisk). The second hairpin is short, containing the SD sequence base paired with a ROSE-like motifCitation23,Citation27 exhibiting a bulged uridine (U90). SD and AUG are highlighted in gray circles, the putative anti-SD sequence is marked by squares. Black encircled/framed Nt are referred to in the quantification in (C). (B) Enzymatic structure probing of avalong-RNAT. In vitro transcribed, 5′labeled avalong RNA was partially digested with RNase T1 (0.002U) and RNase V1 (0.01U) at the indicated temperatures. For the control (C) A. dest was added instead of enzyme. An alkaline ladder (LOH) was loaded for orientation. The asterisk marks the cleavages for the alternative anti-SD (C). Quantification of altered RNase T1 (upper panel) and V1 (lower panel) cleavage intensity between 25–42 °C for selected nucleotides. Band intensities were quantified from (B) using the Alpha Ease software (Alpha Innotech, Biozym). Shown are relative intensities for each Nt position with band intensity at 25 °C set as 1.
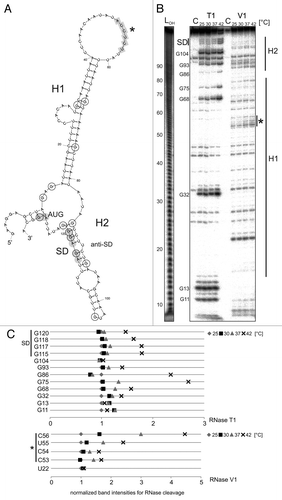
Figure 7. Reporter gene assay with avalong-RNAT WT and variants. (A) Using site-directed mutagenesis, the terminal H1-loop (Nt 31–70), the bulged uridine 90 in the ROSE-like anti-SD was deleted (ΔU90) and two flanking GC pairs were opened by exchanging the cytosines at positions 88 and 91 against adenines (CC88/91AA). Compared with the WT exhibiting a 10-fold increase after heat shock from 30–42 °C, deletion of the top-loop resulted in high β-galactosidase activity at 30 °C and decreased heat-induction after the shift to 42 °C. In contrast, deletion of the predicted bulged U90 provoked low levels of reporter activity both under heat shock and non-heat shock conditions. A loosened structure around the SD led to a high reporter gene expression at 30 °C and 42 °C. The results reflect mean values of a double-measurement; mean standard deviation is given by error bars. (B) Measurement of β-galactosidase activity at 30 °C and 42 °C of avalong-variants harboring a mutated alternative anti-SD sequence. Neither the substitution of C53, C54 by adenines (avalong*2A), nor of the CCUC motif (Nt 53–56, avalong*4A) led to a derepression at low temperatures compared with the wild-type.
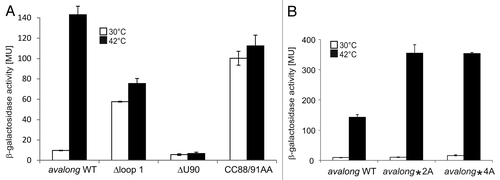
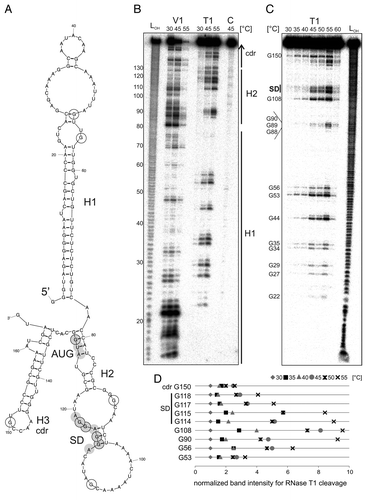
Figure 8. Temperature-dependent structural alterations of the hspA-5′UTR. (A) Model of the T. elongatus hspA-5′UTR and 40 Nt coding region as well as the first two Nt of the blunt end restriction site HpyCH4v according to a secondary structure prediction computed with mfold.Citation86 The structure exhibits two RNA hairpins H1 and H2 in the 5′UTR, the second containing SD region and AUG start codon, as well as a third H3 in the coding region. SD sequence und AUG start codon are highlighted by gray circles. Black encircled Nt are referred to in the quantification in (D). (B) Enzymatic structure probing of the hspA-5′UTR with 40 Nt coding region. In vitro transcribed RNA was 5′labeledCitation81 and subjected to partial digestionCitation82 with RNase V1 (0.001U) and T1 (0.002U) at the indicated temperatures. For the control, (C) A. dest was added instead of enzyme. An alkaline ladder (LOH) was loaded for orientation. Samples were separated on an 8% denaturing polyacrylamide gel. (C) Temperature stability of the hspA RNA structure. Labeled RNA was digested by RNase T1 (0.002U) within a temperature range from 30–60 °C in 5 degree steps. The probing reveals slight accessibility of the SD region for RNase T1 at 45 °C and high susceptibility at 55 °C. Please note that RNase T1 was inactive at 60 °C. (D) Quantification of altered RNase T1 cleavage intensity between 30–55 °C for selected guanines. Band intensities were quantified from (C) using the Alpha Ease software (Alpha Innotech, Biozym). Shown are relative intensities for each Nt position with band intensity at 30 °C set as 1. cdr, coding region.