Figures & data
Table 1. Examples of computational methods for RNA 3D structure modeling that are capable of using experimental restraints
Table 2. Low-resolution experimental methods that generate particularly useful data for computational prediction of RNA 3D structure
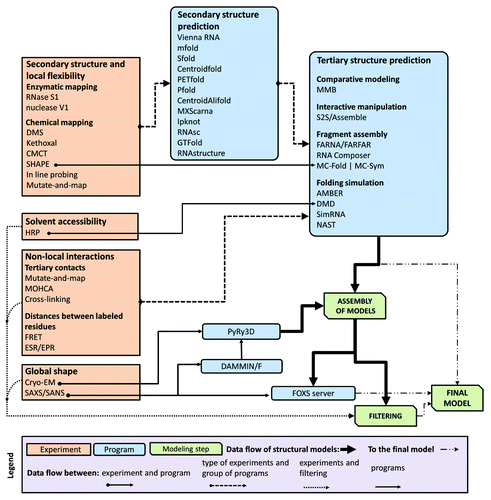
Figure 1. Crystal structure of Escherichia coli 5S rRNA (PDB ID: 3OAS) (A) and computational models predicted with the fragment assembly approach based on structural probingCitation78 (B) and manual modeling based on cryo-EM data of the 50S subunitCitation79 (C).
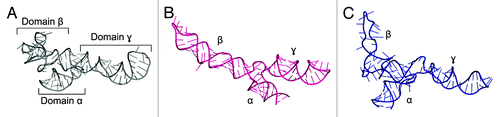
Figure 2. A model of VAI∆TS RNA structure obtained with SimRNA and built into the SAXS reconstruction using PyRy3D.Citation85
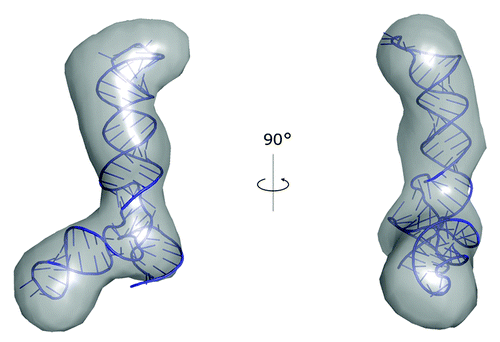
Figure 3. A flowchart describing relations between different types of data, computer programs, and RNA 3D structure modeling strategies.