Figures & data
Figure 1. Co-immunoprecipitation of chromosomally tagged Hfq and bound RNA. Wild-type and C-terminally tagged-Hfq strains (3xFlag or HA) were grown to exponential phase. Cell lysates were subjected to immunoprecipitation using anti-Flag or anti-HA-coupled beads. Cell lysates (input) and immunoprecipitated fractions (anti-Flag and anti-HA) were separated on 12% SDS-PAA gel. (A) Proteins were transferred onto a nitrocellulose membrane and immuno-blotted with anti-Flag or anti-HA antibody or (B) visualized by Silver staining. Bands corresponding to 3xFlag- (*) and HA-Hfq (#) are indicated. (C) Cells with C-terminally tagged Hfq bearing either 3xFlag or HA, hfq deletion and isogenic wild-type cells were grown to exponential (E) and stationary phase (S). Cell lysates were separated on an 8% SDS-PAA gel, transferred onto a nitrocellulose membrane and immuno-blotted with anti-RpoS antibody. (D) Equal amounts of co-immunoprecipitated RNA were analyzed by chip-based capillary electrophoresis.
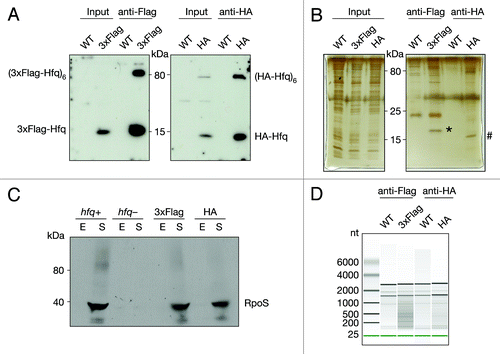
Figure 2. Categorization of Hfq-binding RNAs based on deep sequencing. Hfq-binding RNAs were categorized as (A) full-length RNAs, (B) antisense RNAs, (C) intraRNAs, or (D) intergenic RNAs. Annotated coding features (ORFs or RNAs) are represented as arrows and identified intervals are represented as bars. Dark gray elements indicate Hfq-enrichment, while white elements indicate non-enrichment; light gray indicates that an element can be either Hfq-enriched or non-enriched. The wavy line represents the predicted Hfq-binding RNA. For more details see Materials and Methods.
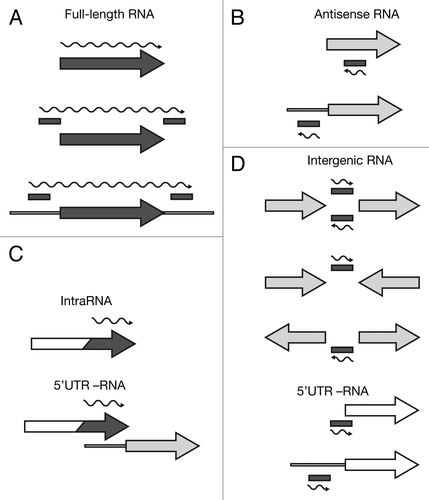
Figure 3. Identification and verification of antisense RNAs binding to Hfq. Deep sequencing results of as-intF and as-gspM (A and C) represented as averaged coverage maps of 3xFlag- and HA total RNAs and 3xFlag- and HA-co-immunoprecipitated RNAs (Hfq co-IP). The genomic strands are shown in blue (+) and red (-). Note that scales for the + and – strand differ. The genomic location is depicted on top and the genomic context is depicted below. The red bar indicates the position of the oligonucleotide probe that was used in the corresponding Northern. The wavy line represents the predicted position of the novel RNA. Northern blot analyses of as-intF and as-gspM (B and D). Hfq-3xFlag total RNA and RNA co-immunoprecipitated with Hfq-3xFlag (left panel) and total RNA isolated from hfq deletion strain (hfq-), corresponding isogenic wild type (hfq+), RNase III mutant (rnc-) and corresponding isogenic wild-type cells (rnc+) (right panel) were fractionated on a denaturing 8% polyacrylamide gel, electro-blotted onto a nylon membrane and hybridized with radioactively labeled oligonucleotide. Note that different amounts of RNA were loaded; 20 μg total RNA and 1 μg co-IP RNA. Ladder sizes in nt are indicated. 5S RNA was used as a loading control. At least two independent experiments were performed and representative data are shown.
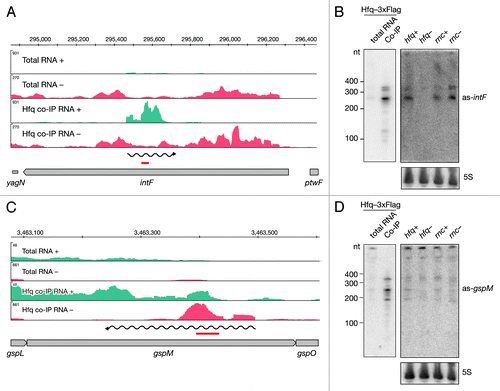
Figure 4. Identification and verification of novel Hfq-binding intragenic RNAs. Deep sequencing and Northern blot analyses of intraRNAs intra-yadD (A and B) and intra-narK (C and D) as described in and .
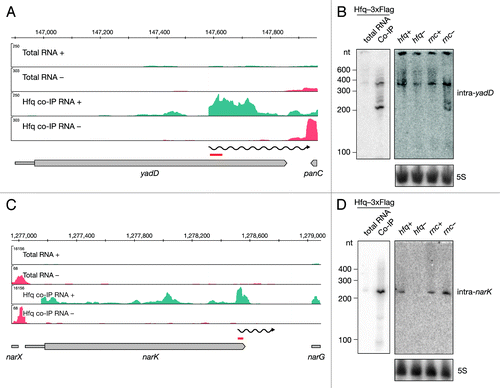
Figure 5. Identification and verification of tRNA and sRNA precursors binding to Hfq. Deep sequencing and Northern blot analyses of Spf (A and B) and the metZWV precursor (C and D) as described in and . Violet and red bars indicate the positions of oligonucleotide probes used in the corresponding Northern blots. 6S RNA was used as a loading control. The black bar indicates the position of the Hfq aptamer reported by Lorenz et al.Citation9
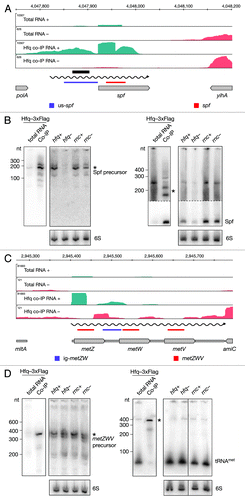
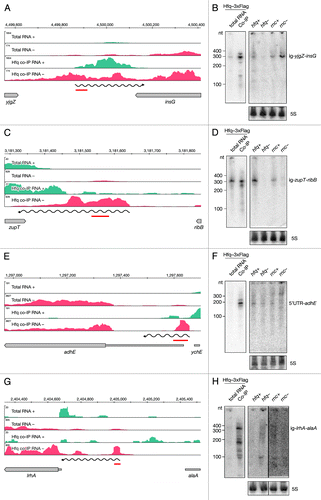
Figure 6. Identification and verification of novel Hfq-binding intergenic RNAs. Deep sequencing and Northern blot analyses of intergenic RNAs yjgZ-insG (A and B), zupT-ribB (C and D), 5′ UTR_adhE (E and F) and lrhA-alaA (G and H) as described in and .