Figures & data
Figure 1. Overview of computational predicted and experimentally identified sRNAs in S. meliloti. On the left, Venn-diagram illustrating the number of potential sRNA encoding regions predicted by del Val et al.,Citation5 Ulve et al.,Citation6 and Valverde et al.Citation7 On the right, Venn-diagram representing sRNA transcripts identified via RNA-seq studies by Schlüter et al.,Citation8 Schlüter et al.,Citation27 and Torres-Quesada et al.Citation28 and a comparison to the 156 predicted candidates. Number in brackets: clustered sRNA genes (at least two experimentally identified neighboring sRNA genes) which were spuriously predicted and summarized as a single sRNA gene region were counted as individual transcription units.
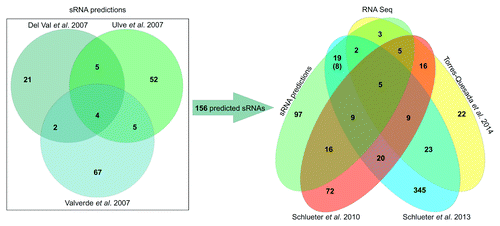
Figure 2. Comparison of two A. tumefaciens dRNA-seq studies. Venn diagram comparing the sRNAs identified by Wilms et al.Citation11 and Lee et al.Citation12
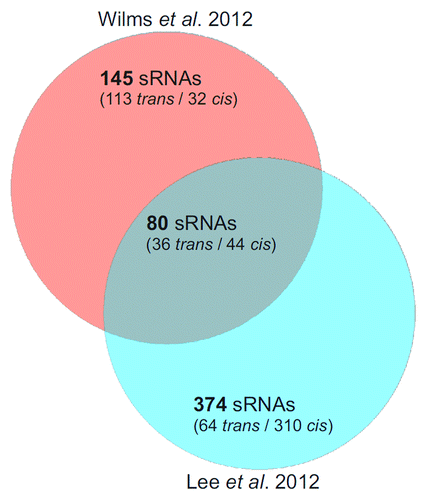
Figure 3. Distribution of trans-sRNAs in the Rhizobiales. The circular plot is based on a simplified phylogenetic tree adopted from Reinkensmeier et al. and del Val et al.Citation34,Citation38 Sequenced strains which harbor at least one sRNA of a defined RNA family model (RFM) are illustrated. Each circular row incorporates all sRNA members of an RFM and shows their assignment to members of the Rhizobiales. The two RFMs separated by the green circle were defined exclusively by del Val et al.Citation38 The RFM nomenclature in brackets indicates the availability of alternative RFMs calculated by del Val et al.Citation38 (not shown here). Grey, chromosomal sRNAs; light blue, sRNAs encoded on pSymB-like replicons; dark-blue, sRNAs encoded on pSymA-like replicons. Multi-colored rows indicate RFMs with several RNA gene homologs per strain/genome: Yellow, single gene; orange, two homologs; pale violet red, three homologs; green, four or more homologs. Species glossary: R. l., Rhizobium leguminosarum; R. e., Rhizobium etli; A. r., Agrobacterium radiobacter; A. t., Agrobacterium tumefaciens; A. v., Agrobacterium vitis; A. sp., Agrobacterium species, S. m., Sinorhizobium meliloti; S. e., Sinorhizobium meliloti; S. f., Sinorhizobium fredii; B. a., Brucella abortus; B. m., Brucella melitensis; B. s., Brucella suis; B. mi., Brucella microti; B. o., Brucella ovis; B. c., Brucella canis; O. a., Ochrobactrum antrophi; M. l., Mesorhizobium loti; M. c., Mesorhizobium cicero; M. sp., Mesorhizobium species; P. l., Parvibaculum lavamentivorans; M. e., Methylobacterium extorquens; M. c., Methylobacterium chloromethanicum; X. a., Xanthobacter autotrophicus; St. n., Starkeya novella; Az. c., Azorhizobium caulinodans; Rh. p., Rhodopseudomonas palustris; N. w., Nitrobacter winogradskyi; Be. i., Beijerinckia indica; Met. s., Methylocella silvestris; Rm. v., Rhodomicrobium vannielii.
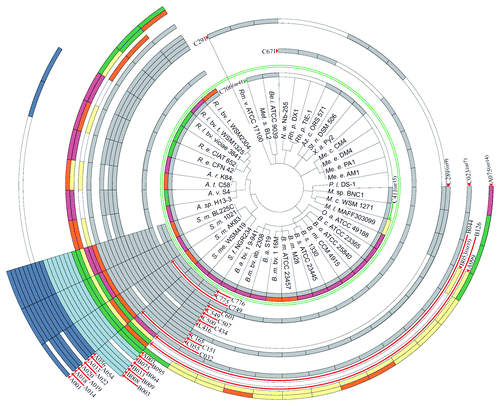
Figure 4. Structural Alignment of AbcR1/2 (SmelC411), SmelA075, and SmelA099. Consensus secondary structures are colored according to the Vienna RNA conservation coloring scheme.Citation110 A. Structural alignment of RFM αr15 (AbcR1/2) defined by del Val et al.Citation38 In yellow, anti-Shine-Dalgarno (aSD) sequences (modules M1 and M2) in single-stranded regions and loop structures. Pale red, Hfq binding site. B. Structural alignment of RFM SmelA075 and SmelA099 defined by Reinkensmeier et al.Citation34
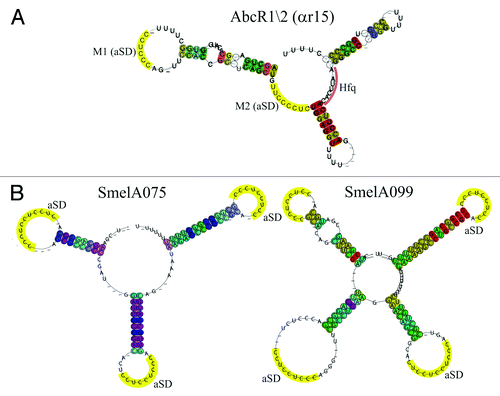
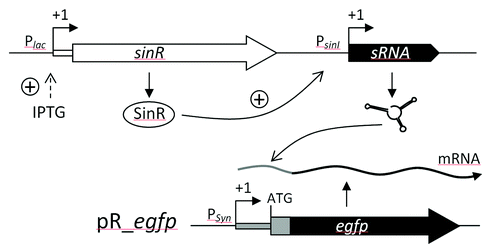
Figure 5. sRNA pulse overexpression construct based on IPTG inducible expression of sinR which activates PsinI-driven sRNA gene expression. The effect of sRNA overproduction is monitored applying a reporter construct based on plasmid pR_egfp.Citation40 PSyn is a constitutive promoter derived from Plac.