Figures & data
Figure 1 Mss116p-promoted splicing of the ai5γ intron in yeast mitochondria. (A) Comparison of the in vivo splicing activity in a wt yeast strain and a mss116-knockout strain. In the presence of Mss116p 70–75% of the cox1 pre-RNA are spliced, while less than 1% of splicing occurs in the absence of Mss116p. (B) Scheme of the Poison Primer Assay used to assess in vivo splicing activities. A primer hybridizes to the 5′ end of the downstream exon and is extended by a Reverse Transcriptase in the presence of ddCTP (and dATP, dGTP, dTTP). The extension therefore stops at the first G in the template, which results in a Primer + 14 nts product for the ligated exons and Primer + 8 nts for the unspliced pre-RNA.
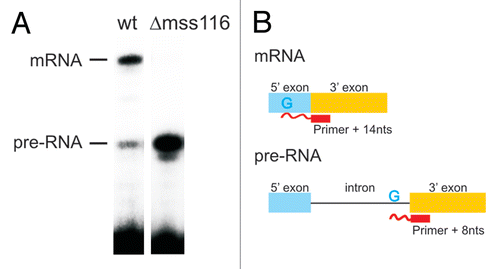
Figure 2 The intracellular structure of the ai5γ intron. (A) Representative primer extension gel showing the in vivo DMS modification pattern of the ai5γ intron - region at the c-c1-c2 three-way junction - in the wt yeast strain. (A and C) denote sequencing lanes. In the - lane natural stops of the Reverse Transcriptase are seen. In the + lane the in vivo DMS pattern is shown. Notably, comparing lanes 3 and 4 reveals the DMS-induced stops of the Reverse Transcriptase and thus accessible residues (N1-A, N3-C). Additional regions of the intron are shown in the subsequent figures ( and ; Suppl. Figs. 2, 3 and 5). (B) Summary map in which residues methylated by DMS are indicated with blue filled circles. The size of the filled circles correlates with the relative modification intensity of individual bases. As the spliced intron form was analyzed, it was not possible to map the structure of domain 6 shown in gray (as it served as primer binding site to map the very 3′ part of the intron). D2 and D4 are shown in Supplementary Figures 2 and 3, respectively.
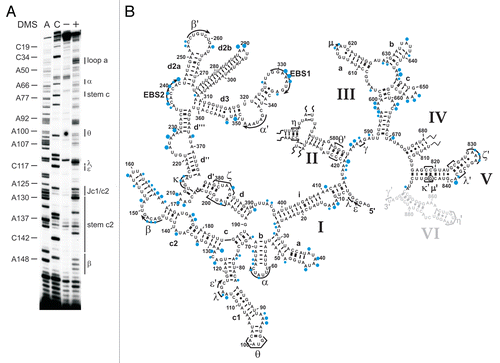
Figure 3 The κ-ζ element depends on Mss116p for folding. Representative gel showing the modification intensity of nucleotides in the 5′ part (A) and 3′ part (B) of the D1 core structure composed of stems d, d′ and d″, in which the κ region, the ζ receptor and the coordination loop are embedded, in the wt and mss116-knockout strain. The arrow heads indicate residues, the accessibility of which changes due to the absence of Mss116p (filled arrow heads represent an increase in accessibility, while open ones highlight bases with reduced accessibility). Strong changes in accessibility are displayed in red (>2-fold); while smaller changes are shown in gray (1.5 to 2-fold). These values were derived from the normalized gel plots (see Suppl. Fig. 4A and B). Lanes are designated as in with the exception that − and + lanes are shown for both the wt and mss116-knockout strain. Comparing lanes 4 and 6 reveals the altered DMS modification pattern and thus conformational changes within the ai5γ intron due to the absence of Mss116p.
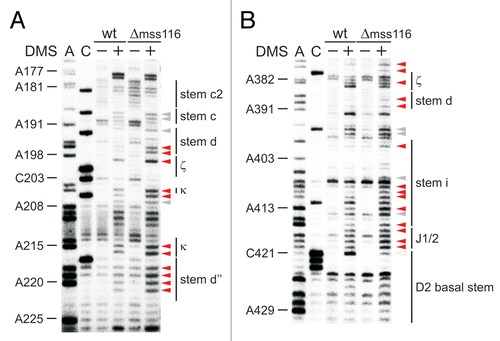
Figure 4 Tertiary structure elements are unfolded in the absence of Mss116p. (A) Representative gel showing the accessibility of nucleotides in the c-c1-c2 three-way junction, which harbors the θ, λ, ε′ and β elements forming long-range tertiary contacts with distinct regions of the intron, in the wt and mss116-knockout strain. Lanes are designated as in ; symbol code as in . The normalized plot derived from this gel is shown (Suppl. Fig. 4C). (B) Representative gel showing the accessibility of residues in stem d3 including the exon binding site 1 (EBS1) and the α′ element, which forms an intra-domain contact with the closing loop of stem b. Lanes are designated as in ; symbol code as in . The normalized plot derived from this gel is shown Supplementary Figure 4D.
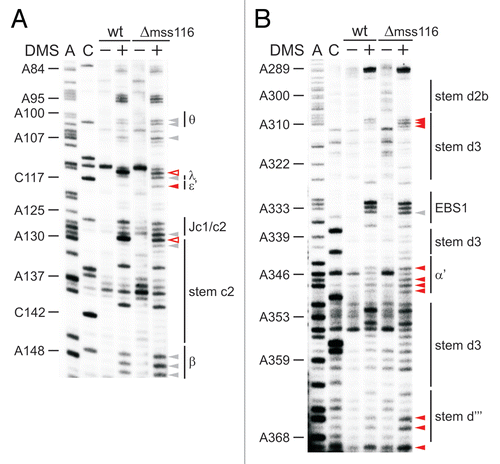
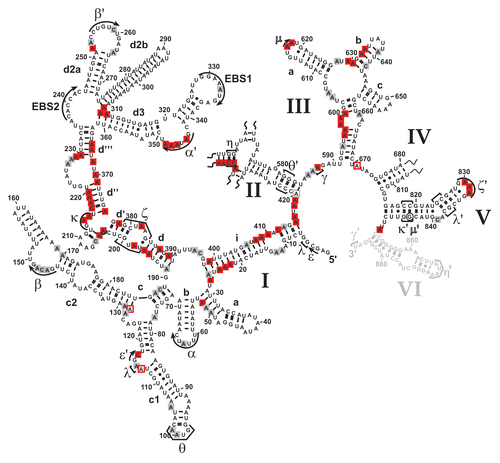
Figure 5 Mss116p-induced conformational changes within the ai5γ intron in vivo. Differential summary map: residues, the modification intensity of which changes in the absence of Mss116p (Δmss116 strain), are highlighted. This map is based on normalized plots (see Suppl. Figs. 4 and 6), which had been derived from the modification gels. The closed squares indicate an increase in accessibility; while open squares represent bases with reduced accessibility in the absence of Mss116p. The (A and C) residues whose modification remains unaltered (i.e., equally modified or protected in both strains) are not highlighted. Color code as in . D2 and D4 are shown in Supplementary Figures 2 and 3, respectively.