Figures & data
Figure 1 Interactions in the EJC cycle. The scheme shows the sequence of either simultaneous or mutually exclusive interactions underlying function of the EJC and its sub-complexes. The EJC is assembled during splicing onto the mRNA. After the mRNP is exported into the cytoplasm to function in translational regulation (ribosome in gray), the EJC is disassembled with the aid of a ribosomal-binding protein, PYM (in orange). The MAGo-Y14 heterodimer (in blue and cyan, respectively) is re-imported into the nucleus without PYM by Imp13 (in yellow). Here, it is released by RanGTP binding (gray/black rectangle) and incorporated into a spliced mRNP with eIF4AIII and BTZ (in pink and red, respectively) in the form of the EJC (other mRNP proteins are shown in gray with different shapes and sizes). mRNP composition varies dynamically as well as in the composition of the periphery of the EJC. UPF3b (in purple) associates to the EJC in the nucleus and travels with the complex to the cytoplasm. The question marks indicate open issues in the cycle: it is unclear whether eIF4AIII and BTZ are imported into the nucleus separately or as a single unit and the putative import receptor(s) has not been identified yet.
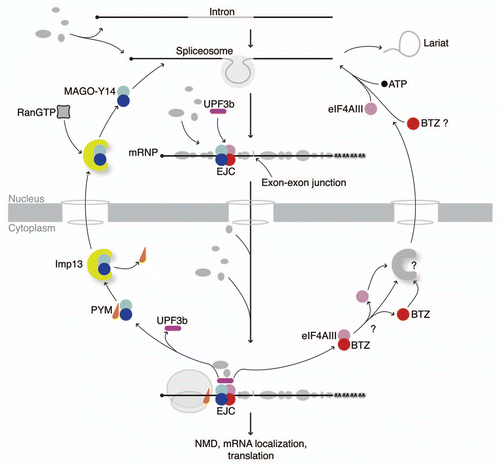
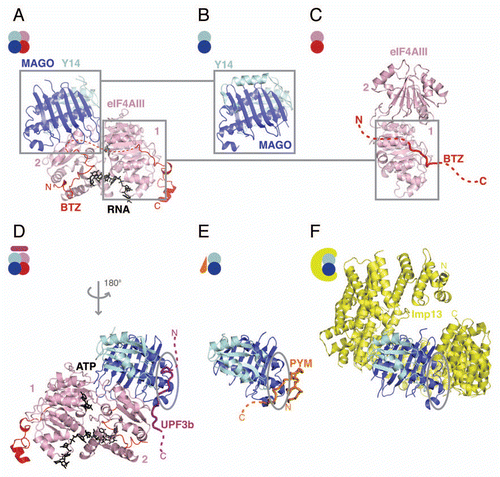
Figure 2 Structures in the EJC cycle. (A) Structure of the human EJC (pdb id. 2J0Q). eIF4AIII is shown in pink and BTZ in red, with ATP and RNA shown in black. The N- and C-terminal domains of eIF4AIII are labeled as 1 and 2, respectively. MAGO is shown in blue and Y14 in cyan. The disordered loop of BTZ is represented as a dashed red line. The N- and C-termini of the protein are labeled. The same color-coding is applied to all the panels and is consistent with the scheme in ; (B) Structure of the MAGO-Y14 heterodimer in isolation from Drosophila (pdb id. 1HL6) in a similar view as in (A); (C) Structure of the eIF4AIII-BTZ binary complex (pdb id. 2J0U in a similar view as in (A). The disordered portions of BTZ that could not be modeled in the structure are shown as a red dashed line; (D) Crystal structure of the EJC-UPF3b complex (pdb id. 2XB2). The complex is shown with a 180° rotation around the y-axis as compared with the orientation of the complex in (A). The UPF3b peptide is rendered as a purple cartoon tube. The dashed line represents parts of the protein that were not modeled in the structure. Highlighted with a gray circle is a conserved patch on MAGO-Y14 that is involved in protein-protein interactions also in (E and F). (E) Structure of the Drosophila MAGO-Y14-PYM complex (pdb id. 2RK8). PYM is rendered as a cartoon tube in orange. The N- and C-termini of the protein are labeled. (F) Structure of the Imp13-MAGO-Y14 complex with Imp13 in yellow (pdb id. 2X1G). The Imp13 N- and C-termini are indicated. This and all other protein structure figures were generated using PyMOL (www.pymol.org).