Figures & data
Figure 1 Schematic presentation of the genomic locations/flanking genes of the sRNA-Xcc1 homologues associated with integrons (A) and transposons/plasmids (B). The black arrow represents sRNA-Xcc1 homologue, the gray arrow represents open reading frame (ORF) and the ORF number and/or the predicted coding product was shown inside or under the arrow. The number on the top of a vertical line indicates the genomic position, and the number above a horizontal line indicates the length of the spacer between the sRNA-Xcc1 homologue and the upstream or downstream genes (or genetic element). The corresponding bacterial strains were shown on the right hand side of the figure. The genomic data were obtained from the NCBI nucleotide database.
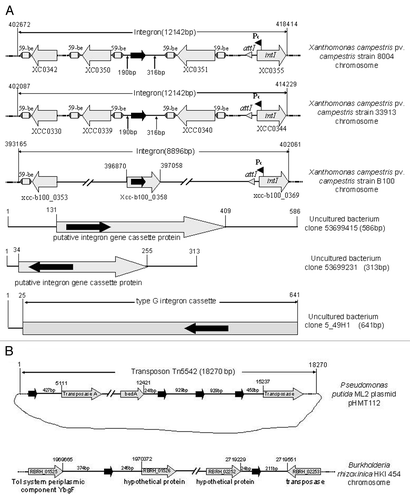
Figure 2 The phylogenetic tree of sRNA-Xcc1 homologs based on multiple alignments. Symbols on the right hand side of the name of the bacterial strain indicate the class of the bacterial species, and the location of the sRNA-Xcc1 homolog.
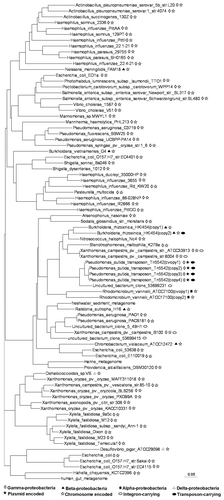
Figure 3 (A) Schematic presentation of the genetic organization of the integron of Xcc and the detailed composition of sRNA-Xcc1 cassette. The blue letters indicates the sRNA-Xcc1 coding sequence and the red letters indicate the 59-be sequence. (B) Detection of the sRNA-Xcc1 level in the wild type strain 8004, the hrpG mutant 8004ΔhrpG and the hrpX mutant 8004ΔhrpX, by using northern Blot.
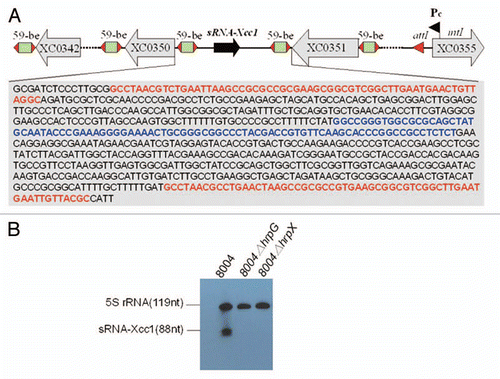
Figure 4 Multiple alignments and a consensus secondary structure model of the sRNA-Xcc1 homologs. Multiple alignments were done using the ClustalW program and the consensus secondary structure was predicted based on the multiple alignment using RNAalifold program. The conserved sequence motif ‘AUACAAnACCC’ was boxed.
