Figures & data
Figure 1 Alternative splicing, a new layer of control of circadian clock gene expression in different eukaryotes. The figure shows that alternative splicing regulates the expression of different components of the central oscillator in Drosophila, Neurospora and Arabidopsis. Exons are shown as boxes, introns as lines, 5′ss are 5′ splicing sites or donors, and arrowheads at the end of exons represent that the gene continues. Black arrows indicate activation and black lines with a bar at the end indicate inhibition. Left upper part, Drosophila melanogaster circadian oscillator's scheme. The cartoon shows the central genes involved in the circadian clock central loops in this organism. Per, period; Tim, timeless; dClk, Drosophila Clock; Cyc, cycle. Left bottom part, per alternative splicing isoforms. Middle upper part, Neurospora crassa circadian oscillator's scheme. l-Frq and s-Frq, long and short isoforms of Frequency respectively, which arise from the two different translation initiation sites (flag AUG); WC, white collar. PU and PD are alternative promoters of frequency. Middle bottom part, frq alternative splicing isoforms depending on the promoter used. Right upper part, Arabidopsis thaliana circadian oscillator's scheme. The cartoon shows the central genes involved in the circadian clock in this organism. TOC1, TIMING OF CAB EXPRESSION 1; LHY, LATE ELONGATED HYPOCOTYL; CCA1, CIRCADIAN CLOCK ASSOCIATED 1, PRR7 and PRR9, PSEUDO RESPONSE REGULATOR 7 and 9, respectively. Right bottom part, PRR9 and CCA1 alternative splicing isoforms.
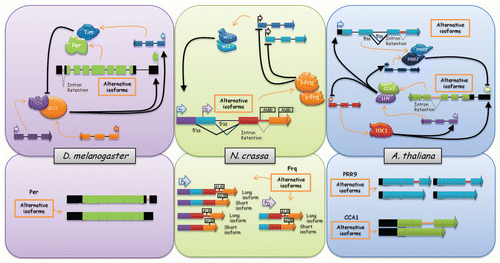
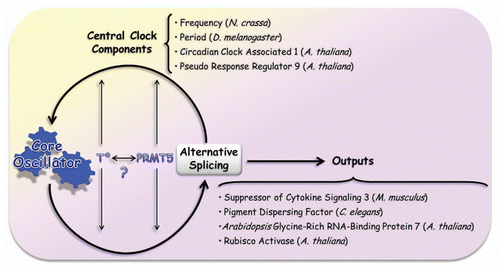
Figure 2 Alternative splicing and its relationship with circadian clocks. The curved arrows indicate that the central core of the circadian clock is known to modulate alternative splicing, allowing a certain isoform to peak in a particular momentCitation29,Citation30 and that, in turn, alternative splicing plays a regulatory role over the central oscillator by modulating pre-mRNA splicing of essential pieces of this mechanism. As explained in the text, these interactions are regulated by temperature and PRMT5 protein (straight and thin arrows). It is still unknown whether there is a connection between those factors. There are several examples of alternative spliced genes which are involved in the modulation of circadian outputs. Among them, we mentioned the case of Suppressor of Cytokine Signaling 3 (SOCS3) in M. musculus,Citation47 Pigment Dispersing Factor (PDF) in C. elegans,Citation48 and Glycine-Rich RNA-Binding Protein 7,Citation49 and Rubisco ActivaseCitation30,Citation50 in A. thaliana. Gene targets of alternative splicing and components of the central core are cited: Frequency (N. crassa),Citation22 Period (D. melanogaster),Citation19,Citation20 Circadian Clock Associated 1,Citation7 and Pseudo Response Regulator 9 Citation30 (A. thaliana).