Figures & data
Figure 1 Bru protein and binding motifs. (A) Organization of Bru protein. The structure is shown schematically, to scale, with the three RRM RNA binding domains indicated. The subdomains of Bru used for selections, RRM1+2 and RRM3+, are shown. (B) Graphical representations of preferred binding motifs identified by in vitro selections. The height of each stack represents the information content at each nucleotide of the motif in bits. At top left is the predominant motif identified from the Bru selection. The other motifs were identified from the RRM3+ selection. (C) comparison of the BRe consensus sequence and similar motifs from the selections. (D) The 3′ UTRs of the indicated mRNAs are shown schematically, with motifs from the aptamer selections indicated. The full height bars are perfect matches to the motifs, while the half height bars have a single mismatch. The top five 3′ UTRs are of Bru target mRNAs, while the bottom two 3′ UTRs are from other mRNAs not known to be regulated by Bru.
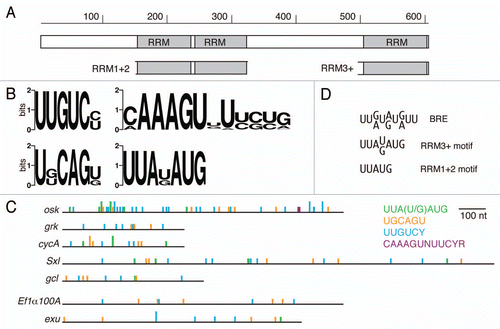
Figure 2 Bru binding to Anti-Bru aptamers. (A) Results of filter binding assays with recombinant Bru (0.9–477 nM) and anti-Bru aptamers. (B) Aptamer sequences (excluding the sequences from the cloning vector that are common to all) are shown, with KD values obtained from the data of part A. Motifs identified from the in vitro selections are shaded. The predominant UUGUCY motif from the Bru selection is shaded brown, with only perfect copies indicated. All of the anti-Bru aptamers have at least one copy of a singly-mismatched version of the motif (these are not shaded), while the control RNA obtained from the pool prior to selection (r0.3) has none. Examples of the motifs from the RRM3+ selection that also appeared in the Bru selection are shaded in green and red. Instances of the 5 tetranucleotides that appear most frequently in the anti-Bru aptamers (UUGU, UUUU, UGUC, UUUG and UGUU; Sup. Table 4) are shaded in yellow (except where the sequences are within another shaded region). The proportion of U, which is highly enriched in the Bru selection, is given for each RNA.
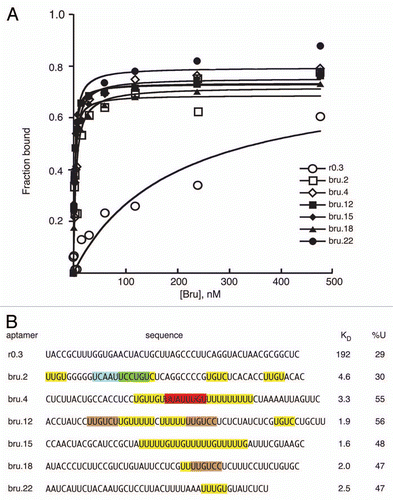
Figure 3 Translational repression by candidate regulatory sites. (A) schematic diagram of reporter mRNAs with the variable region indicated by a filled box. For the SV40 reporter, the variable region is only SV40 sequences. For the remaining reporter mRNAs the variable region has four copies of a candidate Bru regulatory site embedded in SV40 sequences. (B–H) examples of GFP levels in stage 10A egg chambers expressing GFP reporter transgenes with the matα4-GAL-VP16 driver. All confocal images were taken on the same day at the same settings. The scale bar represents 75 µm. (B) control GFP transgene with no anti-Bru aptamer binding motifs. The remaining image parts are for GFP transgenes with the Bru binding motifs indicated in the figure and described below. (C) UGUUUUAUAUGU is from the osk AB region, and consists of a BRE-like motif adjacent to a short U/G rich motif (like those from the RRM1+2 selection). (D) CAAUUUUAUAUGU is from the cycA 3′UTR, and consists of a BRE-like sequence adjacent to a short C/A rich motif (like those from the RRM1+2 selection). (E) UCAAUUGCAGU is from the cycA 3′ UTR, and consists of a copy of the UGCAGU motif (from the RRM3+ selection) adjacent to a short C/A rich motif (like those from the RRM1+2 selection). (F) UGUUUGUAGU is from the grk 3′UTR, and consists of the UGCAGU motif (from the RRM3+ selection but with a single mismatch) adjacent to a short U/G rich motif (like those from the RRM1+2 selection). (G) UUGUCc is the type II Bru binding site, which appears three times in the AB and C regions of the osk 3′UTR. (H) UAAAGUCUUCUA is from the osk C region, and is a type III Bru binding site with a single mismatch relative to the longest aptamer motif from the RRM3+ selection. (I) Relative GFP levels in the nurse cell cytoplasm of stage 9/10 egg chambers for each of the reporter transgenes. GFP levels (obtained from 45 measurements for each transgene) were normalized to the RNA levels (from 3 measurements for each transgene). Transgene RNA levels were normalized relative to rp49 RNA levels. All of the reporter transgene mRNAs with candidate regulatory sites show reductions in GFP levels that are significantly lower than for the control (p < 0.0001 by the Tukey-Kramer method).
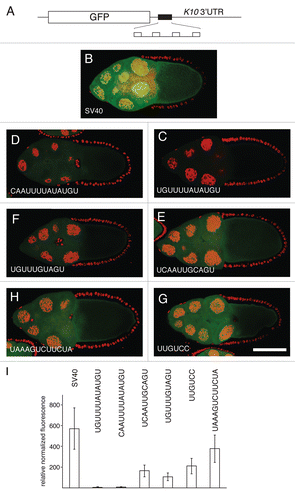
Figure 4 Translational repression of reporter mRNAs requires Bru. (A–G) The pairs of parts show GFP expressed from a reporter mRNA in aret−/+ ovaries (A–G, left) or aret−/aret− ovaries (A′–G′, right). The identity of the Bru regulatory sites is shown below, with the relative increase in GFP level from mutation of aret indicated beneath the mutant parts. The scale bar represents 50 µm. In all cases the driver was nosGAL4VP16, which is active at early stages of oogenesis (the aret mutant ovaries arrest oogenesis and do not progress to the stage shown in Figure). The transgenes are the same as those in Figure, with four copies of a particular Bru binding motif as indicated.
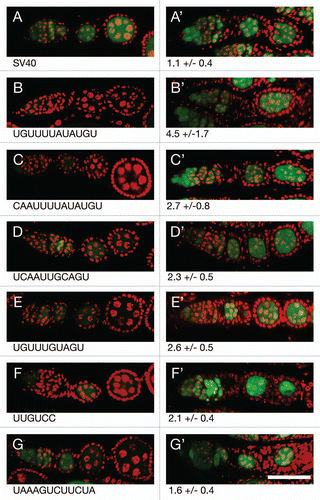
Figure 5 Bru binding to regulatory sites. (A) UV crosslinking assay with ovarian protein and RNAs bearing four copies of the Bru binding sites embedded in SV40 sequences. The RNA probes are indicated at top by the identity of the binding site. The SV40 probe is SV40 RNA alone, and the osk AB probe is the AB region of the osk 3′ UTR. (B) competition binding assay. Crosslinking assays of the type shown in (A) were repeated, with or without the presence of unlabeled competitor RNAs. The competitors are osk AB RNA, or the same RNA (all-) with point mutations in BREs and type II Bru binding sites.Citation17 exposures of the different rows are not equivalent, but were chosen to have similar binding signals in the absence of competitor. (C) Filter binding assay with recombinant Bru (1.26–322 nM) and the RNAs used in (A and B).
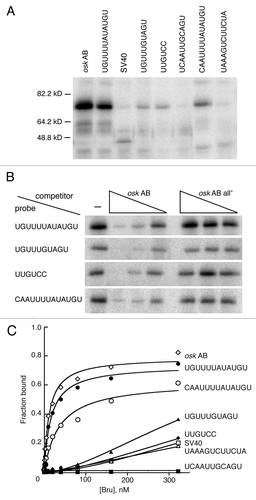
Figure 6 Models for Bru binding. (A) combinatorial binding: different RNA binding domains of Bru interact with extended binding sites in the same substrate RNA. (B) Independent binding: different RNA binding domains interact with different substrate RNAs. (C) Bridging binding: different molecules of Bru bind to the same substrate RNA. For simplicity, the possible third Bru RNA binding domain identified from the Bru selection is not shown, as its position in the protein is unknown.
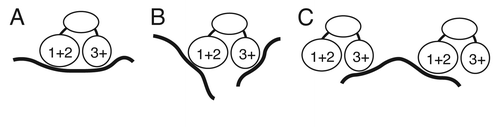
Table 1 Highly enriched tetranucleotides in the RRM1+2 aptamers
Table 2 Nucleotide composition of aptamers and osk 3′ UTR regions