Figures & data
Figure 1 Identification of pri/pre-miRNA loop mutations that potentiate cel-let-7 activity and partially rescue defective mature let-7 biogenesis of cel-let-7 miRNAs in human cells. (A) Schematic diagrams depict the pre-miRNA sequences and structures of the wild-type cel-let-7 gene and loop mutants. (B) Seed-dependent repression of target reporter by the cel-let-7 and loop mutants. Average results of at least six independent trials ± S.D. are shown. Statistical significance was determined by an unpaired two-tailed Student's t test. (C) Expression of pre- and mature miRNA made from wild-type and loop mutant cel-let-7 genes was determined by quantitative northern blot. Representative results of four northern blot analyses from independent transfections are shown. Blots were also probed for U6 small nuclear RNA as a loading control. (D & E) LP4 mutations in cel-let-7 result in increases in pre- (n=2) and mature let-7 (n=3 to 6) with correct 5′ ends. The 5′ ends of (D) mature let-7 and (E) pre-let-7 made from the wild-type cel-let-7 and cel-let-7_LP4 genes were determined by primer extension analyses.
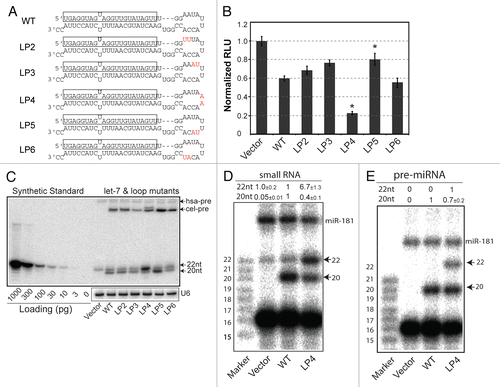
Figure 2 Loop mutations that inhibit biogenesis of full-length mature let-7. (A) Schematic diagrams depicting the pre-miRNA sequences and structures of cel-let-7_LP4 and loop mutants. (B) A northern blot analysis representative of three experiments shows pre-miRNA and mature miRNA expression from cel-let-7-LP4 and loop mutants. (C) The 5′ ends of mature let-7 miRNAs made from the type cel-let-7 and pri-let-7 loop mutants were determined by primer extension analyses (n=6). (D) The 5′ ends of pre-let-7 miRNAs made from the type cel-let-7 and pri-let-7 loop mutants were determined by primer extension analyses (n=2).
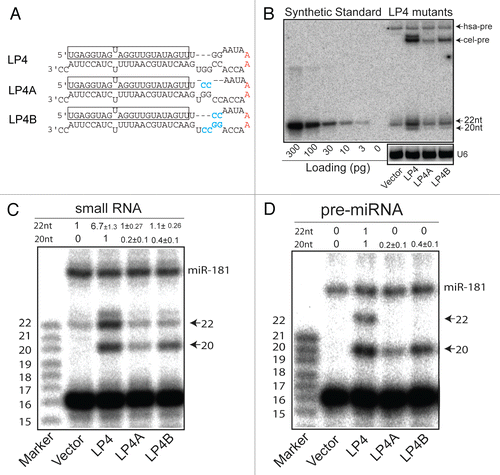
Figure 3 The activities of cel-let-7_LP4 and loop mutants in target repression correlate with the formation of specific pri-let-7 RNAs and target complexes. (A) Repression of luciferase reporter expression by cel-let-7_LP4 and loop mutants. Representative results of at least six independent trials (± S.D.) are shown (*, p<0.0001 except as indicated). (B) Formation of pri-let-7 RNAs and target complexes predicts the activity of cel-let-7 and loop mutants in target repression. Lin-41_LCS reporter mRNAs were tagged with S1 aptamers and used to pull-down associated pri-let-7 RNA. S1-tagged reporter mRNAs with the Lin41-LCS_sm UTR or without the Lin41-LCS UTR were used as negative controls. The ratios of wild-type to mutant pri-let-7 RNA and corresponding S1-tagged target RNAs with or without the wild-type lin-41-LCS UTR or with a seed mutant lin-41-LCS UTR in the pull-down RNA samples were determined by using qPCR analyses (n=4, ±S.D., *, p< 0.001). (C & D) Standard curve-based qPCR was carried out to determine the average copy numbers of (C) pri-let-7 and (D) mature let-7 per target in the pull-down samples (n=4, ±S.D.). (E) The effects of cel-let-7-wt and cel-let-7-LP4 expression on the levels of target mRNA were determined by qPCR analyses (n=4, ±S.D., *, p< 0.001).
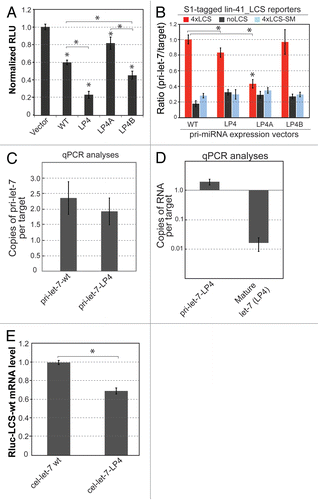
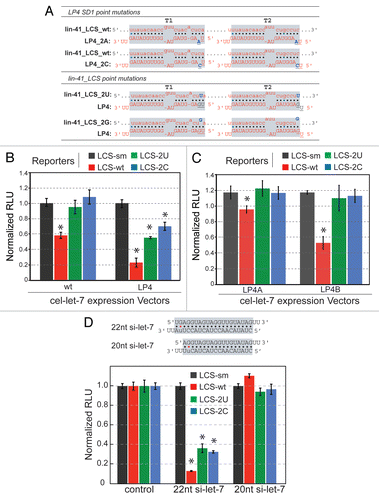
Figure 4 Repression activity by cel-let-7 and loop mutants may be controlled by both SD1-sensitive and SD1-insensitive components. (A) Schematic diagrams showing the predicted base parings between let-7 and modified lin-41_LCS. Altered nucleotides are indicated in blue, missing as underlined. (B & C) Repression of mutant lin-41_LCS reporters that abolish SD1 pairing by (B) wild-type cel-let-7 and cel-let-7-LP4 and (C) double loop mutant cel-let-7-LP4 genes. (D) siRNA duplexes of 20 base pairs (truncated let-7) and 22 base pairs (full-length mature let-7) were transfected into cells expressing mutant lin-41_LCS reporters that abolish SD1 pairing and reporter activity was measured. Representative results of at least six independent trials (± S.D.) are shown in panels B, C, and D (*, p< 0.01).