Figures & data
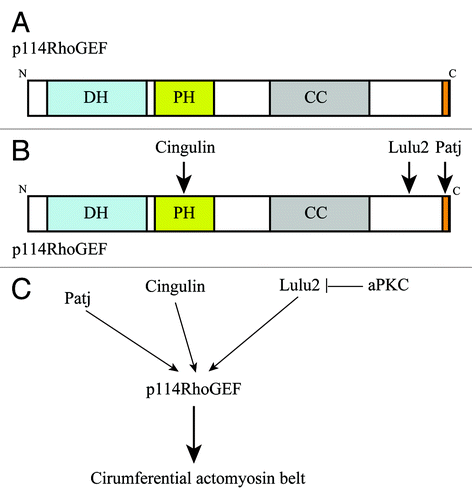
Figure 1. The circumferential actomyosin belt regulates apical constriction. (A) The circumferential actomyosin belt is positioned along apical cell-cell junctions (tight and adherens junctions) in polarized epithelial cells. (B) Myosin II-dependent contractile forces of the circumferential actomyosin belt induce apical constriction in epithelial cells.
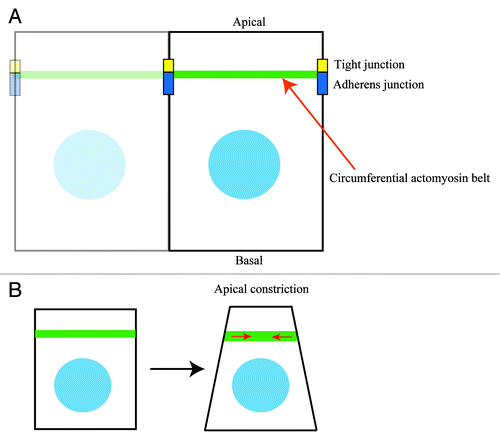
Figure 2. Lulus commonly have FERM and FERM-adjacent domains. (A) Both Lulu1 and Lulu2 have two alternative spliced transcripts that share FERM and FERM-adjacent domains in their N-terminal portion. The C-terminal portions of the isoforms do not obvious homology among them. (B) FERM and FERM-adjacent domains are conserved not only between Lulu1 and Lulu2, but also across species. The numbers indicate similarities or identities of the domains compared with Lulu1 (left or right, respectively).
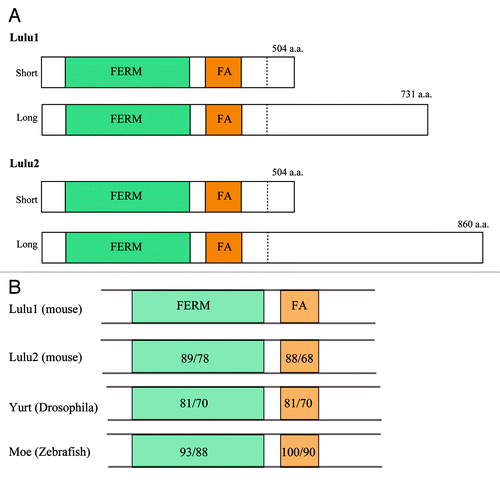
Figure 3. p114RhoGEF is activated by Lulu2. (A) p114RhoGEF has Dbl homology and pleckstrin homology (PH) domains, which are necessary for its catalytic activity, followed by a coiled-coil region and a PDZ domain-binding motif in its C-terminal tail. (B) p114RhoGEF interacts with cingulin via the PH domain, Lulu2 via C-terminal portion, and Patj via the PDZ domain-binding motif in its C-terminal tail. (C) Cingulin and Patj anchor p114RhoGEF at apical cell-cell boundaries. Lulu2 interacts with and activates p114RhoGEF. aPKC inhibits Lulu2. This molecular system regulates the circumferential actomyosin belt.