Figures & data
Figure 1 Graphical summary of the evolution of the CDC25 Homology Domain as described in van Dam et al. For each class of RasGEFs we have traced back their perceived and reconstructed time of origin. For the mammalian RasGEFs we reconstructed the time of duplications to specific points in eukaryotic evolution. Note the differential loss of ancestral RasGEFs between animals and fungi. All gene names of fungal RasGEFs are from orthologs in S. cerevisiae unless specified otherwise. The evolutionary time scale is based on Douzery et al. but note that molecular dating is highly inaccurateCitation51 and that these dates are therefore approximate at best.
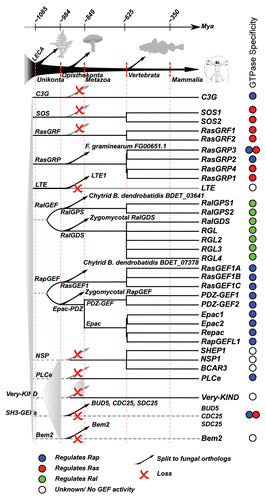
Figure 2 Representation of the phylogenetic tree of RapGAP domain containing proteins. We observe four clades, each of whicht represent a single ancestral gene in LECA . RalGAPB and RalGAPA each represent a single ancestral gene in LECA but cluster together indicating a common ancestral gene preceding LECA . The species present in each clade is depicted as a colored barcode. The differences in sequence between the Rap specific GAP sequences and the Rheb/Ral specific GAP sequences is well defined (100% bootstrap support) and a possible root may therefore lie between these two groups. *In the Rap specific clade we also observe GARNL3, a putative RapGAP, but it cellular function has not yet been reported. #It is unknown if RalGAPB harbors any GAP activity.
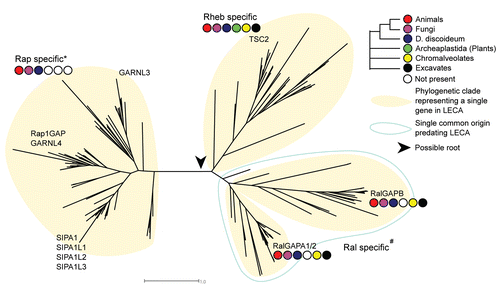
Figure 3 Simplified representation of the phylogenetic tree of RasGAP containing protein sequences. Gene names from human and S. cerevisiae are depicted as representative genes. Representative domain architectures for each clade are depicted in the tree and the five clades representing a single ancestral gene in or before LECA are numbered. The GAP1 and IQGAP clades from a combined clade in which a domain architecture is shared that predates LECA . The tree depicted has been modified to reflect proper grouping based on domain architecture: the clade containing RASA-like genes in D. discoideum, chromalveolates and N. gruberi is found between clusters 2 and (3,6) in the ML tree, see Figure S2.
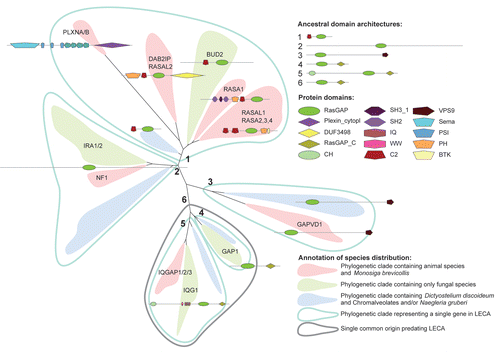
Figure 4 Evolutionary timeline of the Ras-like subfamily members. Many duplication events occurred in the common ancestor of the metazoa and vertebrates. The LECA likely contained multiple ancestral Ras-like GTPases although it is impossible to resolve how many there exactly were. Confident branches are depicted in black. Grey lines indicate possible alternative interpretations but are supported by circumstantial evidence. Gray dotted lines are based on a strict interpretation of the position of these GTPases in the phylogenetic tree but are otherwise unsupported and may originate from a more recent ancestral gene in the Unikont or Opisthokont ancestor. Fungal gene names are based on the orthologous genes in the yeast S. cerevisiae. The evolutionary time scale is based on Douzery et al. but note that molecular dating is highly inaccurateCitation51 and that these dates are therefore approximate at best.
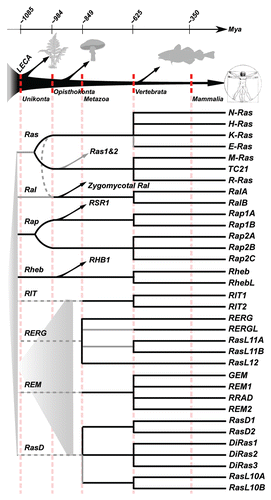
Figure 5 Evolutionary reconstruction of the Ras, Ral, Rap and Rheb GTPases based on co-evolution with their regulatory domains. (A) Matrix describing for each GTPase subtype which regulatory domain is active. (B–O) All scenarios possible for the order of duplications that gave rise to Ras, Rap, Ral and Rheb. For each scenario we reconstructed the order of invention and loss of regulation by the regulatory domains to fit the observed regulation of the GTPases as depicted in (A). Parts that exhibit the minimal number of events needed to fit the matrix in (A), (B) (4) and (C, D, H, M and N) (5 events) are marked.
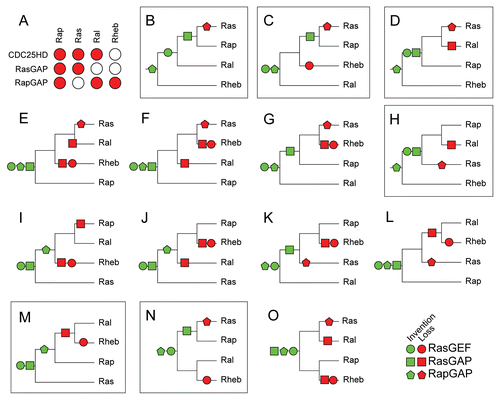
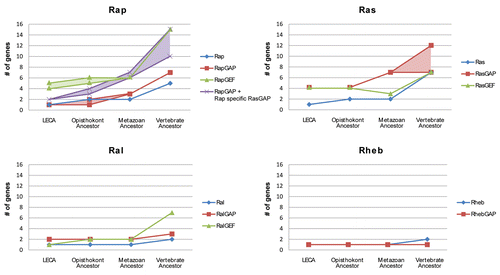
Figure 6 Expansion of Ras-like GTPase subtypes and their respective GEFs and GAPs (Ras-like regulatory system) in time leading up to mammalian organisms. We incorporated upper and lower estimates for GEFs and GAPs based on how the phylogenetic trees can be interpreted. The difference between higher and lower estimates for RapGAPs and RasGAPs is mainly caused by the RASA1 RasGAP protein family as GTPase specificity shifts within this family (see main text). The Rap GTPase regulatory system display a many to one GEF regulation network. This schema is maintained throughout eukaryotic evolution leading up to vertebrates. However with the inclusion of the C2-RasGAPs, the GAP regulation of Rap shows a similar trend. In contrast, the Ras regulatory system displays a many to one GAP regulation, while maintaining a relatively equal amount of Ras GTPase specific GEFs. The difference in the rate of expansion of the regulatory proteins for the Ras and Rap GTPase subtype indicates that there is a fundamental difference between the regulatory networks of Ras and Rap. Where Rap relies on multiple GEF and GAP proteins for its signaling diversification, Ras seems to rely mostly on its GAPs. The Ral and Rheb GTPases display a relatively compact regulatory system although in the vertebrates the number of RalGEFs expands significantly.