Figures & data
Figure 1. Mechanical forces applied on centrosomes. Centrosome positioning is maintained by coordinated microtubule pulling forces and cortical actin pushing centripetal flow. During division, enhanced actomyosin contractility in the periphery of the opposite pole drags adjacent astral microtubules to drive the separation of duplicated centrosomes to opposing poles. Figure adapted from both Burakov and Rosenblatt.Citation4,Citation5 Centrosome (gray), nucleus (pink), recycling compartment (orange), endoplasmic reticulum (green), Golgi (magenta).
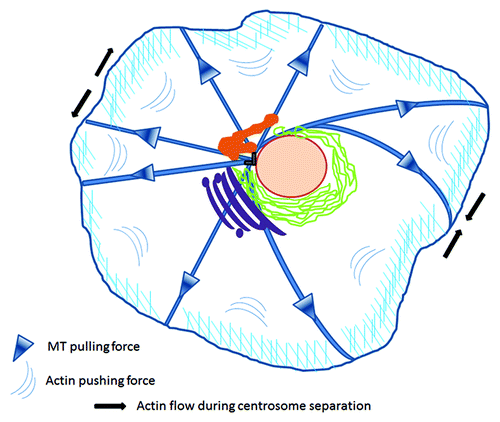
Figure 2. RhoD encircles centrosomes. Preference to one of the two centrin positive couples. HUVE cells were infected with myc-RhoDWT or myc-RhoDG26V and transfected with GFP-Centrin. Twenty four-hours post transfection/infection cells were processed for GFP-centrin, myc-RhoDWT or G26V (red-TRITC) and endogenous α-tubulin (blue-CY5). Bar = 10 μm. Image taken from Kyrkou.Citation32
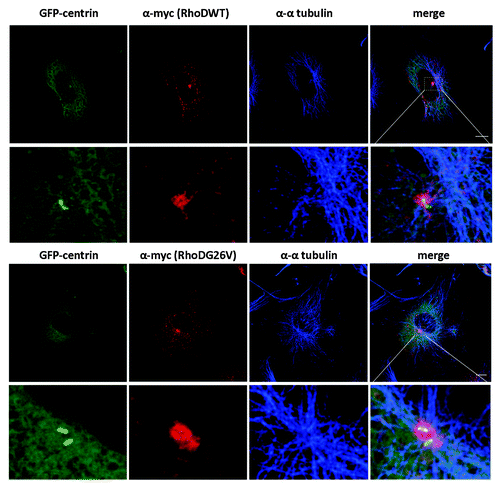
Figure 3. RhoDWT does not colocalize with PCM1-positive centrosome granules. HUVE cells were transfected with myc-RhoDWT for 24 h. Myc-RhoDWT protein was detected with α-myc/Alexa α-mouse 488 and centrosome granules via α-PCM1/ATTO α-rabbit 425. The upper panel is a confocal image. The boxed region in the upper image is shown in the lower panel and is a STED image. Bar = 10 μm.
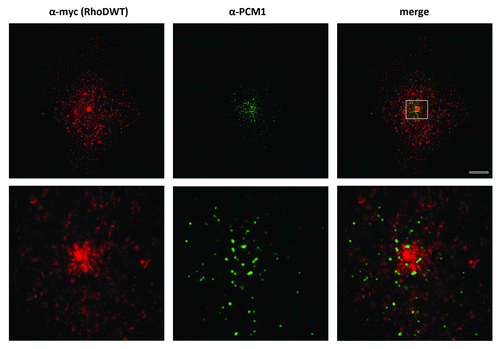
Figure 4. RhoDWT-positive endosomes show partial overlap with Rab11-positive (recycling) endosomes. HUVE cells were co-transfected with myc-RhoDWT and GFP-Rab11WT for 24 h. GFP fluorescence was quenched by fixation in methanol for 20 min at −20°C. Myc-RhoD was detected with α-myc/ATTO-rabbit 425 and Rab11 with α-GFP/Alexa α-mouse 488. The upper panel is a confocal image. The boxed regions in the upper merged image are shown in the central and lower panels and are STED images. The central panels show the colocalization of Rab11 and RhoD in the perinuclear recycling compartment, while the lower panels show the colocalization on peripheral vesicles. Bar = 5 μm. Quantitation with MotionTracking Software yield approximately 56% overlap of RhoD-positive vesicles vs. Rab11-positive endosomes (and 32% of Rab11/RhoD). This is based on total colocalization, while in the perinuclear recycling compartment the colocalization is significantly higher.
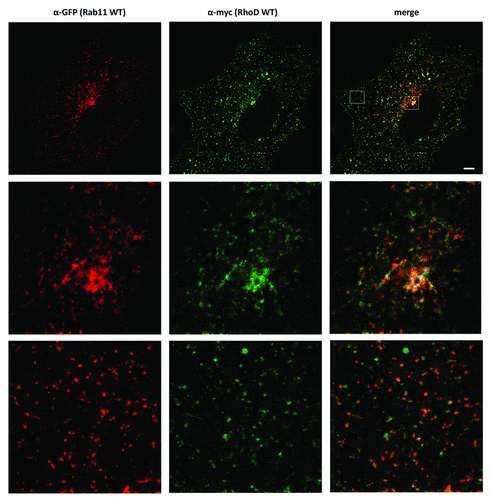
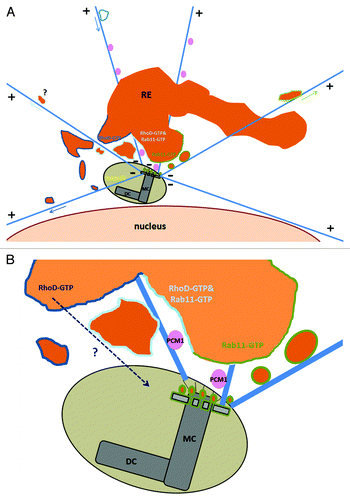
Figure 5. Crosstalk between the recycling compartment and the centrosome. (A) RhoD-positive and/or Rab11 positive vesicles of the recycling compartment surround the centrosome. A distinct population of endosomes (Microtubule driven trafficking in arrow) are either moving from the recycling area (or maybe the periphery) toward the centrosome or emanating from this pericentriolar area to the cell’s periphery. (B) Mother centriole appendages are decorated with recycling endosomes and membrane-free Rab11-GTP.Citation29 RhoD-GTP trafficking inside the centrosome structure is under investigation. Illustrated areas of RhoD-GTP in dark blue lines, Rab11-GTP in green lines, RhoD-GTP and Rab11-GTP colocalization in cyan, PCM1 granules in pink, Rab6c in yellow. MC, mother centriole; DC, daughter centriole.