Figures & data
Figure 1. Phylogenetic tree of human Rho family GAPs. Partial protein sequences (~100 aa) of the GAP domain of 65 human Rho/Rac GAPs were aligned with ClustalW. Reconstruction of the unrooted tree was performed with distance matrix analysis on Mobyle portal in the following order: PROTDIST (Categories model) followed by FITCH (Global rearrangements).
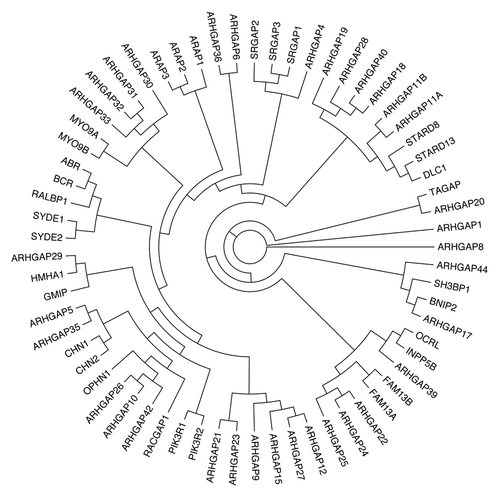
Figure 2. Domain structure of human Rho/Rac GAPs. Abbreviations for domains are as follows: A, alanine-rich; BAR, Bin/amphiphysin/Rvs; C1, cysteine-rich phorbol ester binding; C2, calcium-dependent lipid binding; CC, coiled-coil; CRAL-TRIO, cellular retinaldehyde and TRIO domain; FCH, Fes/CIP4 homology; FF, contains two conserved phenylalanine residues; G, glutamine-rich; IQ, calmodulin-binding motif; Myhl2, myosin head-like 2; MyTH4, myosin tail homology 4; P, proline-rich; PDZ, protein binding site; PH, pleckstrin homology; PX, Phox homology; RA, Ras association (RalGDS/AF-6) domain; S, serine-rich; S/T kinase, serine/threonine kinase; SAM, sterile α motif; SH2, Src Homology 2; SH3, SRC Homology; 3START, StAR-related lipid transfer; WW, two highly conserved tryptophans (in purple). Arrowheads with numbers indicate the position of the catalytic arginine residue in the RhoGAP domain. SH3-binding domain (in blue).
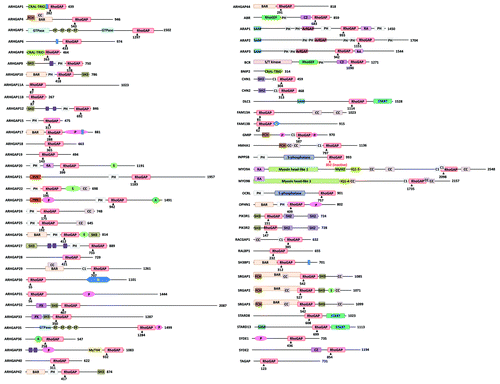
Table 1. Expression profile of human Rho family GAPs based on EST database
Table 2. Expression profile of human Rho family GAPs based on EST tumor database
Figure 3. Comparative analysis of healthy and tumor tissues from EST database. We investigated 53 human Rho family GAPs and 12 equivalent tissues from both healthy and tumor databases. (A) shows the healthy tissues, and (B) shows the tumor tissues. The transcript per million (TPM) values are indicated with color-coded scale. Blue color correlates with the low TPM values, which show the low expression frequency of GAPs. Warm colors are proportional to higher expression frequencies.
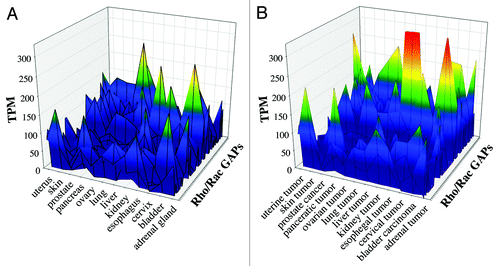
Table 3. Expression profile of human Rho family GAPs based on microarray database
Figure 4. Correlation between two parallel neutrophil samples from microarray data. GSM90844 and GSM90843 samples were analyzed by linear regression. R square is indicated. Pearson Correlation Coefficient = 0.987, p = 6.921E-036. Spearman Correlation Coefficient = 0.913, p = 0.0000002.
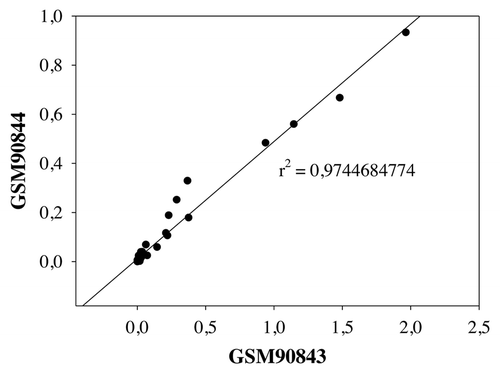
Table 4. Correlation between EST and microarray data
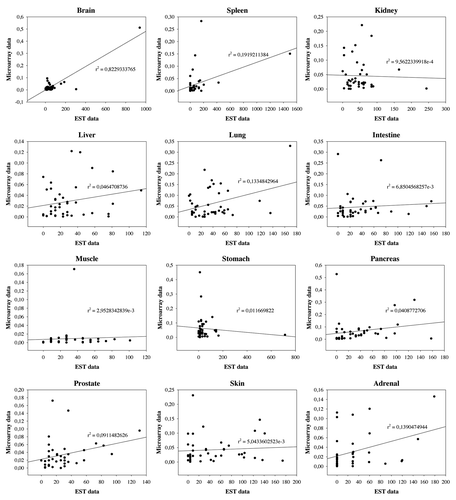
Figure 5. Correaltion between EST and microarray data. We analyzed 12 equivalent tissue types and 41 GAPs from both databases. R square calculated with linear regression is indicated. Correlation coefficients and p values are found in .