Figures & data
Figure 5. Influence of added initiation factors on the translation of the pGEMP/C mRNA. Above the bar graph is a representation of the pGEMP/C mRNA. The bar graph shows the relative levels of the long, medium and short forms of the reporter protein observed in the presence of the added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor. C’ is an in frame extension of C while P is expressed from a reading frame different from C’ and C.
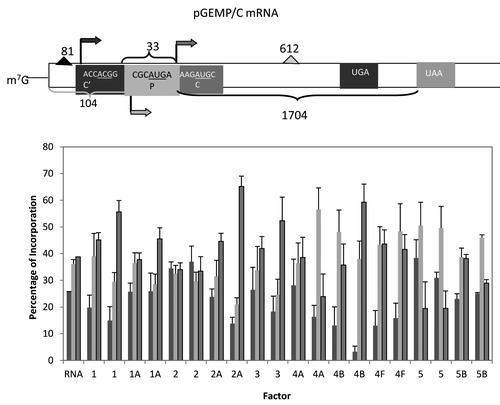
Figure 1. Summary of the optimal mRNA concentrations and reaction times. As described in results, optimal mRNA levels and reaction times for in vitro translation were determined independently and are shown in the figure. Determination of the optimal conditions was based upon hot TCA precipatable [35S]methionine with aliquots taken for the utilization of 0.2, 0.4, 0.6, 0.8 and 1.0 μg of added mRNA or at 0, 10, 20, 30, 40, 60 and 80 min. when an optimal amount of mRNA had been determined.
![Figure 1. Summary of the optimal mRNA concentrations and reaction times. As described in results, optimal mRNA levels and reaction times for in vitro translation were determined independently and are shown in the figure. Determination of the optimal conditions was based upon hot TCA precipatable [35S]methionine with aliquots taken for the utilization of 0.2, 0.4, 0.6, 0.8 and 1.0 μg of added mRNA or at 0, 10, 20, 30, 40, 60 and 80 min. when an optimal amount of mRNA had been determined.](/cms/asset/b59bacc9-b524-427e-8e0b-4bf6c902dd95/ktrs_a_10924419_f0002.gif)
Figure 2. Influence of added initiation factors on the translation of the rGB456 mRNA. Protein synthesis was performed as described in Methods. Panel A shows the total hot TCA precipitable radioactivity obtained in the presence of no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition). For simplicity, the eIF designation is not included in front of the number for each factor. Panel B shows the relative amount of the long (initiated at the first AUG) and the short (initiated at the second AUG) form of the reporter protein (% of the 2 forms such that the long form + the short form = 100%). The relative amount of each protein was determined as described by Laemmli,Citation32 followed by resolution of the two protein bands by SDS PAGE. Dried gels were exposed to X-ray film and then quantitation of the bands was performed by use of a phosphoImager followed by analysis using Imagequant. Above the bar graph is a cartoon representation of the m7G capped mRNA used.
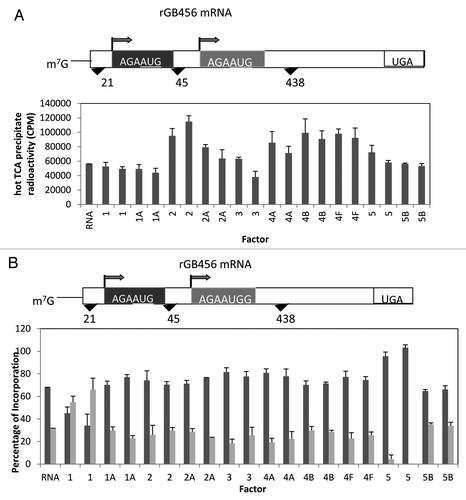
Figure 3. Influence of added initiation factors on the translation of the T7CAT34 mRNA. At the top of the figure is a representation of the T7CAT34 mRNA and below is the relative amount of the long and short form of the reporter protein made in the presence of added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor.
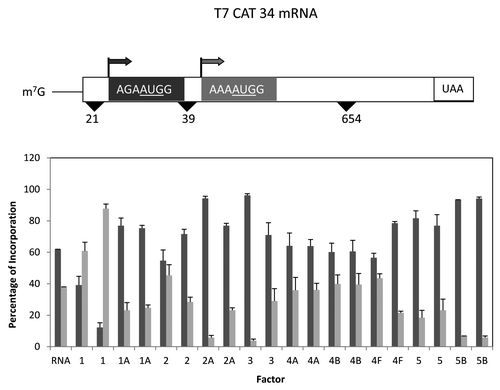
Figure 4. Influence of protein synthesis inhibitors on expression from the rGB456 and T7CAT34 mRNAs. Panel A – Protein synthesis was performed using the rGB456 mRNA as described in with the addition of protein synthesis inhibitors which included: 100 μM m7GTP, 122 nM mp56, 180 nM hp56, 600 pg of poly(I:C) or 0.6 μg of Pdcd4. For each inhibitor, the reaction mixture was incubated with the inhibitor for 15 min. at 30°C prior to the start of the reaction by the addition of mRNA. The control reaction (RNA) was also pre-incubated followed by the addition of the mRNA. Levels of inhibition observed ranged from 40 to 70% except for m7GTP where little inhibition was observed. Shown are the relative amounts of the long form and short forms of the protein made. Panel B – The same analysis as in panel A was performed with the T7CAT34 mRNA.
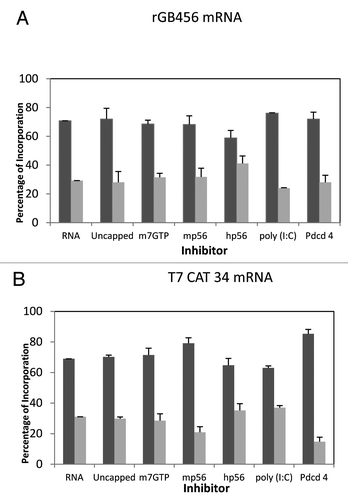
Figure 6. Influence of added initiation factors on the translation of the Pim2 mRNA. Above the bar graph is a representation of the Pim2 mRNA. The bar graph shows the relative levels of the long, medium and short forms of the reporter protein observed in the presence of the added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor.
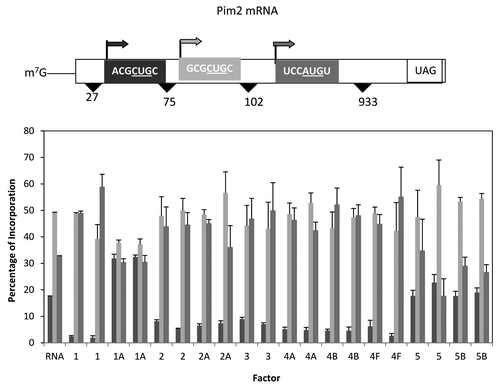
Figure 7. Influence of added initiation factors on the translation of the cMYCCATP2 mRNA. Above the bar graph is a representation of the cMYCCATP2 mRNA. The bar graph depicts the relative levels of the long and short forms of the reporter protein observed in the presence of the added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor.
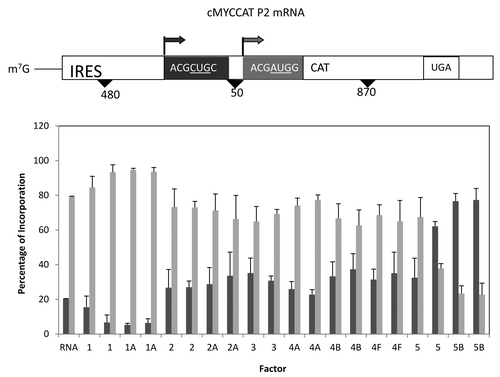
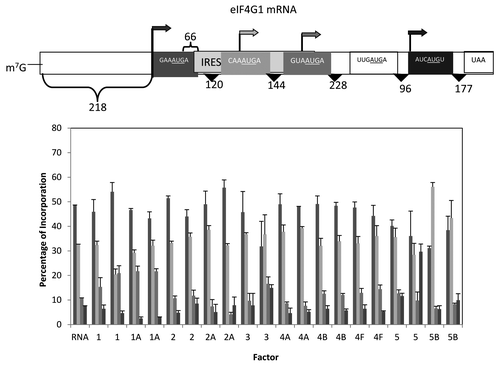
Figure 8. Influence of added initiation factors on the translation of the eIF4G1 mRNA. Above the bar graph is a representation of the eIF4G mRNA. The bar graph shows the relative levels of the four forms of the reporter protein (as indicated by the arrows) observed in the presence of the added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor. No initiation was observed for the possible start codon UUGAUGA.