Figures & data
Figure 1. Transcription coupled repair pathway. When the elongating RNA polymerase reaches a DNA lesion on the template strand, it becomes stalled. At this point, some backtracking events may occur (the 3′-end of the nascent RNA is extruded from the catalytic site). Because a stalled RNA polymerase masks the damage from other proteins (e.g., UvrA and UvrB, see below), it needs to be displaced. This task is achieved by the Mfd (TRCF). Mfd dissociates the RNA polymerase and the nascent transcript, and recruits the UvrAB complex. Then, UvrB-DNA is forms the “pre-incision complex” allowing the recruitment of UvrC, which double-incises the DNA molecule. Finally, UvrD removes the damaged nucleotide sequence and PolI synthesizes a new cDNA, which will be ligated by LigA. Note that the precise mechanism of this repair pathway is still not fully understood and several important questions need to be answered. Among them: does Mfd interact with the RNA polymerase and UvrA at the same time? What is responsible for Mfd conformational changes that allow the “UvrA interacting domain” to be exposed?
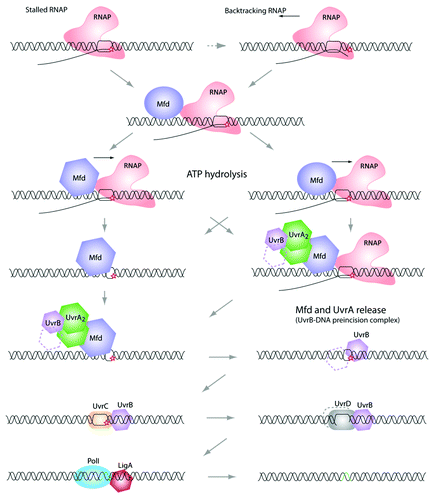
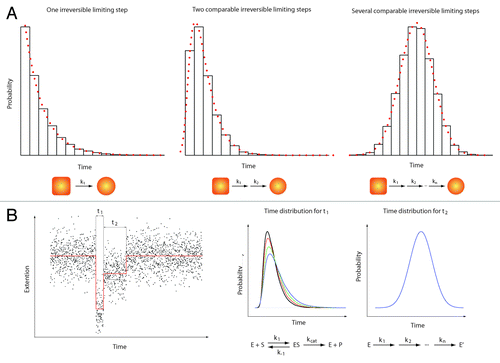
Figure 2. Single molecule enzymology. The most obvious feature of biochemical processes is their stochastic nature. This implies that any biochemical kinetics usually studied in bulk experiments on a large ensemble of molecules can also be cast in terms of probabilities.Citation39 Here, we give a quick overview of the information contained in such statistical distributions. (A) Three examples of statistical distributions (experimental histograms and corresponding probability density functions) obtained for typical bio-chemical processes. From left to right: 1) Single exponential distribution characteristic of one single limiting step with a rate k1. 2) Two-dimensional exponential characteristic of two rate-limiting steps with rate k1 and k2 respectively. 3) Gaussian-like distribution characteristic of several limiting steps with comparable rates (from k1 to kn with k1 ≈k2 ≈… ≈kn). In a typical experiment, hundreds of events (i.e. hundred of single-molecule traces) are needed to build such histograms and get a reliable description of the bio-chemical reactions. (B) Left panel: A theoretical single-molecule extension vs. time trace (e.g DNA extension vs. time trace). This example shows two different events (different DNA extensions) with two different characteristic times t1 and t2. Right panel: The time distribution for t1 (that would be obtained from different molecules) shows a two-dimensional profile. Changing the substrate concentration (e.g., ATP) dramatically affects the shape of the distribution (different colors corresponding to four different substrate concentrations). This behavior is typical of a Michaelis-Menten kinetic.Citation40 In contrast, the distribution of time t2 shows a Gaussian-like behavior that does not depend on the substrate concentration. Such a distribution could be obtained when the transition from two enzymatic forms (say E and E’) occurs through a large number of irreversible rate-limiting steps.