Figures & data
Figure 1. A 9 nt RNA + NTP length gives very strong NTP stabilization of the post-translocation state of the RNAP TEC. (A) Nucleotide scaffolds for pre- and post-translocated TECs (PRE and PST). Template DNA strand (TDS) is blue; non-template DNA strand (NDS) is green; RNA is red. The NTP substrate (red) is in stick representation. Mg2+ is magenta. The closed trigger loop (TL) is yellow. β’ H936 is cyan. The image was derived from PDB 205JCitation5 and drawn using Visual Molecular Dynamics.Citation38 (B) Schematic of experiments for downstream border exo III mapping at TEC-G8 and TEC-A9. * indicates a 32P radiolabel; # indicates a sulfur for oxygen substitution in the TDS to block exo III (orange) digestion. Arrows indicate the upstream to downstream direction of transcription. The positions of the i and i+1 sites are indicated for pre- and post-translocated TECs. At 40 mM KCl, exo III digestion is blocked primarily at the i+18 position. At higher KCl and/or lower pH, digestion can be slowed at i+19 and i+18 (see below). As in panel A, the TDS is blue, the NDS is green and the RNA is red. The TEC bubble is indicated in outlined letters and pink shading. (C) Effects of NTPs (100 μM ATP or CTP) on chain-terminated 3′dG8 and 3′dA9 TECs. Exo III reaction times are in seconds (s). (D) Translocation of G7, G8 and A9 TECs (no chain termination). KCl is 40 mM; pH is 7.9. Backtracked (BTR), pre- (PRE) and post-translocated (PST) TECs are indicated.
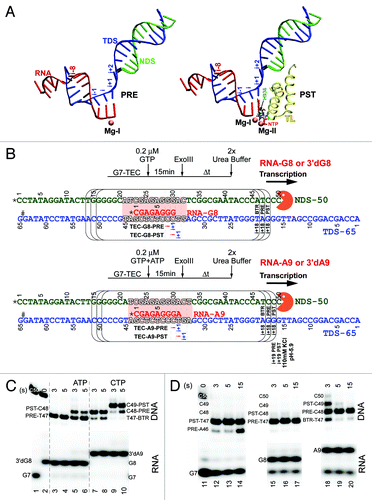
Figure 3. Exo III mapping to obtain a ranking of GTP analogs (400 μM) to stabilize forward RNAP TEC translocation at 40 (upper panel) and 150 mM KCl (lower panel). PPi was at 1 mM. pH is at 7.9.
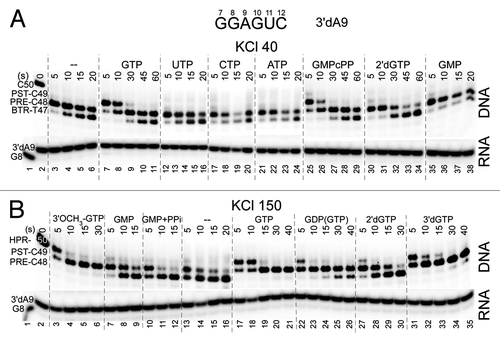
Figure 4. Exo III mapping to obtain a ranking of CTP analogs (400 μM) to stabilize forward RNAP TEC translocation at 40 mM KCl and pH 7.9.
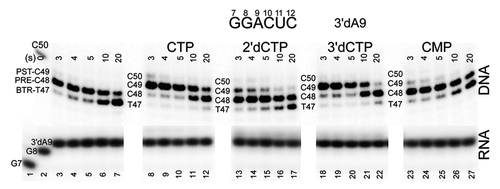
Figure 5. A natural 3′-OH RNA more stably maintains the post-translocation register of RNAP than 3′-H2 and 3′-OCH3 3′ ends, demonstrating the specificity of the exo III mapping assay for the RNAP TEC. GTPαS appears to stabilize the forward translocation state of the TEC slightly more strongly than GTP. GTP and GTPαS were added at 400 μM. The assay was at 40 mM KCl and pH 7.9.
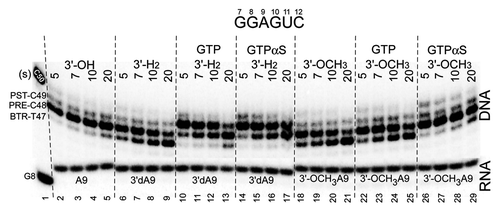
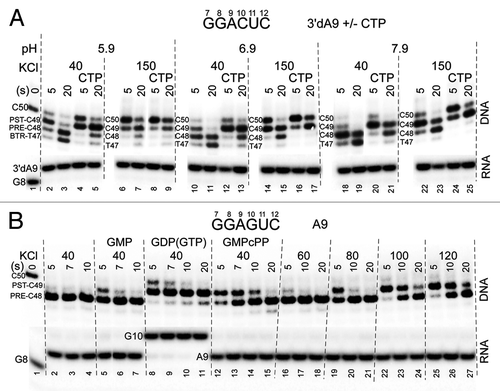
Figure 2. RNAP stalls in the post-translocation register. Forward translocation stability in the absence and presence of NTPs is strongly stimulated by increasing salt and decreasing pH. (A) Exo III mapping of 3′dA9 TECs +/− cognate CTP (400 μM) pH = 5.9, 6.9 and 7.9 and KCl = 40 and 150 mM. The scaffold is as in . (B) Exo III mapping of 3′dA9 TECs +/− cognate GTP or analogs (400 μM). Reactions are shown at KCl = 40, 60, 80, 100, 120 mM (scaffold as in but with a TDS-65 with 34C specifying GTP substrate and complementary 32G NDS-50 at RNA G10). GDP is < 2% contaminated with GTP, which is sufficient to extend A9 to G10 (lanes 8–11). RNA sequences for the two templates are shown.