Figures & data
Figure 1 Identification of optimal co-culturing conditions with a hemolytic strain of S. aureus. (A) Confluent monolayers of HCEC were co-cultured with the hemolytic strain RN6390 using different multiplicities of infection (MOIs). MOI target values were 20, 50, 75 and 100, but the actual experimental value fell within the range indicated. Epithelial cell viability was evaluated by trypan-blue staining, and reported as % viable cells based on total cells counted. (B) Hemolytic activity and bacterial growth of RN6390 and ALC135 cultures, initiated with MOI = 20. The results reflect the activity of bacterial-free culture supernatants on human erythrocyte and are reported as % of total hemolysis. In (B), solid lines represent bacterial growth and dashes indicate hemolytic activity. These data represent the average values ± SD based on two or more independent experiments performed in triplicate. * Significantly different from ACL135-treated group based on the Student's t-test (p value < 0.05).
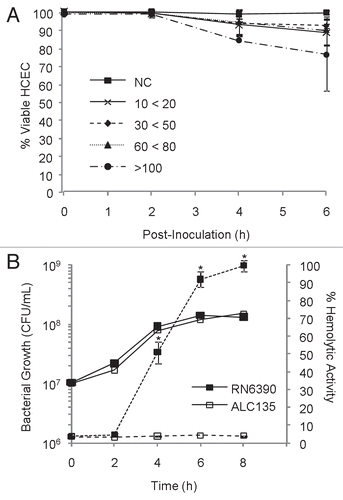
Figure 2 Hierarchial clustering of transcripts produced by HCEC in response to S. aureus RN6390 and the agr−/sar− mutant ALC135. Each column within the graphs represents a single sample, and each row represents a given probe set. The probe sets, shown here, were screened for present detection calls in at least one group with t-test p values ≤0.05. These probe set were further screened for significant differences between treatment groups using an ANOVA analysis with p-value ≤0.01. The intensities of probe sets were considered significantly different between groups if fold-difference ≥2 and t-test p value ≤0.05. Signal values for all probe sets are log2 transformed and normalized by global scaling. Green represents probe set with lower-than-mean levels of transcript, black represent transcripts with mean levels, and red represent higher-than-mean levels of transcript.
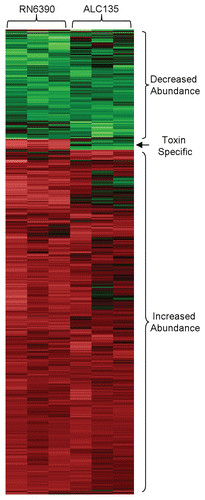
Figure 3 Inflammatory mediators measured in HCEC culture supernatants following co-culture with S. aureus for 6 h at an MOI of 20. (A) Impact of S. aureus on HCEC cytokine and chemokine production. (B) Impact of S. aureus on IFNγ expression in primary HCE and HCLE cultures. Black bars represent media controls. Light and dark grey bars denote co-cultures with strain RN6390 or ALC135, respectively. These data represent the average ± SD of two independent experiments performed in triplicate. *Significantly different from the untreated group based on the Student t-test (p value <0.05). δSignificant difference from RN6390-treated group (p value <0.05).
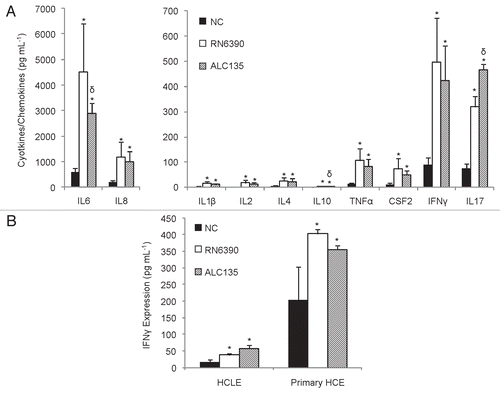
Figure 4 Human corneal epithelial cells secret CCL20 in response to S. aureus exposure but not L. lactis. Supernatants from corneal epithelial and bacterial co-cultures were evaluated for CCL20 concentrations by ELISA. The results are reported as fold change relative to the uninfected control and represented as average values ± SD. (A) HCEC cultures treated S. aureus or L. lactis for 6 h, based on five independent experiments performed in triplicate. (B) Primary HCE cultures treated with S. aureus for 8 h that were sampled at regular time intervals. Data shown based on three independent experiments performed in triplicate. *Significantly different from untreated group based on Student t-test (p value <0.05). δSignificantly different from toxigenic S. aureus strain RN6390 (p-values <0.05).
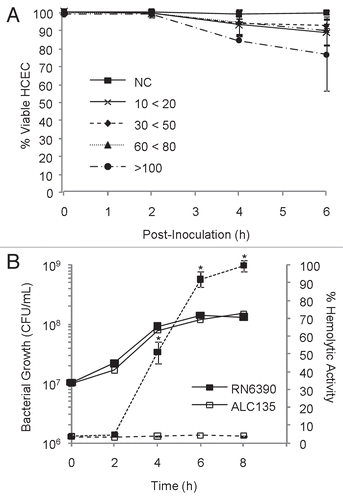
Figure 5 CCL20 production from various sources of corneal epithelial cells upon stimulation with TLR2, TLR4, NOD2 agonists. Confluent monolayers of corneal epithelial cells were stimulated for 12 h with Pam3Cys (10 ug/ml), LPS (10 ug/ml) or MDP (10 ug/ml). Culture supernatants were assessed for CCL20 secretion by ELISA. (A) HCEC cultures (B) Primary HCE cultures and (C) HCLE cultures. The results are representative and graphs display average values ± SD. (A and B) were performed twice and (C) once in triplicate. *Significantly different from untreated group based on student t-test (p value <0.05) and a minimum 2-fold change.
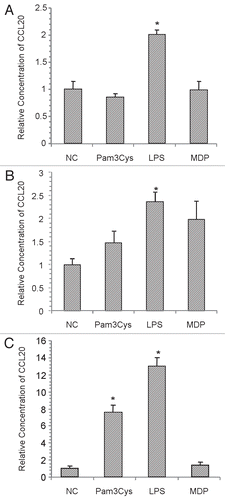
Table 1 Greatest changes in HCECs transcript abundance as the result of exposure to S. aureus
Table 2 Canonical pathways affected by S. aureus
Table 3 Relative abundance of HCEC transcripts determined by realtime PCR and microarray hybridization
Table 4 Transcript in HCEC cultures influenced by S. aureus toxins
Table 5 List of Taqman gene expression assays used to measure relative transcript abundance by RT-PCR as described in materials and methods section