Figures & data
Figure 1 Construction of MP98(CDA2) deletion in strain JEC 34 (serotype D). PC R was used to fuse the promoter region of MP98 (PMP98) and its terminator (TMP98) with the ORF of URA5 to generate a cassette for gene replacement, as shown in (A). Primers are designated in lower case (a–h) and their sequences are given in . Domains of the MP98(CDA2) protein (signal peptide, catalytic and Serine/Threonine-rich) are boxed with “Y” used to identify putative N-glycosylation sites.Citation28 As shown in (B), the strategy to confirm gene replacement for Ura+ transformants was by PC R. Genomic DNAs were used as templates with five sets of primers (g–n, ) to generate products (A–E). Shown below the maps is an electrophoretogram comparing PC R products from wild-type control DNA (1) with a JEC34 Ura+ transformant (2). Each of the five PC R products of the transformant were indicative of gene replacement and a new genotype (mp98::URA5).
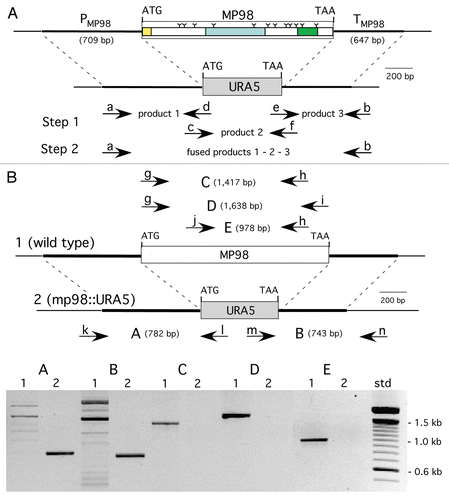
Figure 2 Strain deleted of MP98(CDA2) lacks MP98 protein. Supernatant of a cell homogenate of untransformed strain JEC34 was compared to that of the Ura+ transformant with genotype mp98(cda2)::URA5 ascribed by PCR analysis in . Unstimulated sample had no protein added. Values are the average of four measurements. This bioassay incorporated the specificity of the T-cell hybridoma P1D6 in recognizing an epitope on the MP98(CDA2) protein; recognition activates IL-2 secretion.
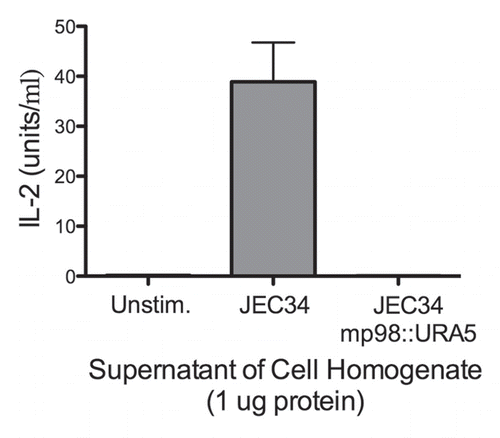
Figure 3 mAb 18F2 is limited to strain JEC21. Immunofluorescence using anti-MP98 MAb 18F2 with serotype D and serotype A, C. neoformans strains. MP98 is located in a punctate pattern in B3501 and near the cell wall in cap67 (top parts). The cda1::URA5 mutant shows an accumulation of MP98 protein in the emerging bud. The negative control, mp98(cda2)::URA5 mutant shows only background fluorescence. Parental strains KN99a and KN99α, mutant cda2Δ and the cda1Δcda2Δcda3Δfpd1Δ quadruple mutant each display only background fluorescence (lower parts). Scale bar is 5 µm.
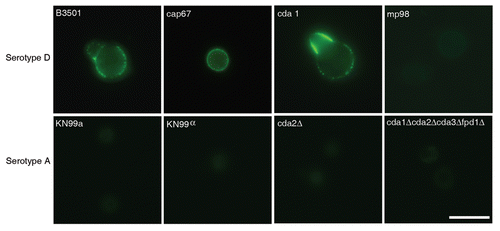
Figure 4 Capsules from B3501 and KN99α had similar permeability to dextran. A dextran penetration assay was performed using tetramethylrhodamine-labeled dextran of molecular weights 10, 40 and 70 kDa. The penetration of dextrans into the capsule was similar for both strains.
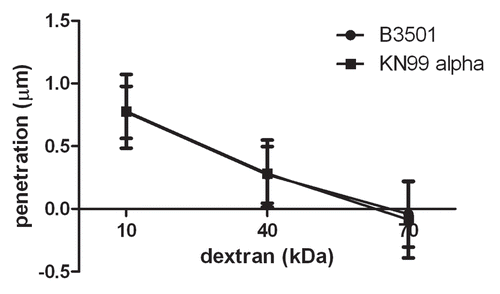
Figure 5 Alignment of MP98 protein sequences encoded by H99, JEC 21 and WM 276 (Serotype B) C. neoformans strains. Sequence highlighted in yellow is for the MP98 protein encoded by the JEC21 allele that was expressed in E. coli and used for generating mAbs. The green areas are putative N-linked glycosylation sites. The red areas are not N-glycosylated in protein encoded by other alleles. The gray areas are possible sites of O-glycosylation differences among protein sequences.

Figure 6 Reconstruction of capsular components. 3D confocal reconstructions from Z-series stacks of B3501 and cap67 cells. Green: GalXM, red: GXM, blue: MP98. Scale bar is 5 µm.
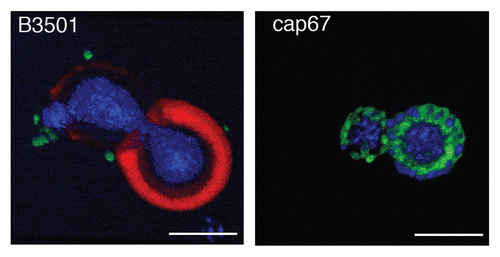
Figure 7 Colocalization studies of MP98 and other mannoproteins using mAb 18F2 and Con A lectin respectively. C. neoformans strain B3501 parts (A–F) and MP98 mutant strain parts (G–I). Left parts are C. neoformans treated with mAb 18F2 (green), middle parts Con A (red) and right parts are the merged images. In B3501 (parts A–F) Con A stains the mannoproteins of cell wall and mannoproteins found on the capsular edge, the mAb 18F2 specifically recognizes and binds to MP98. Parts (A–C) also show that MP98 is mostly localized next to the cell wall during budding. Parts (D–F) shows that 18F2 localizes to bud scars. Parts (G–I) shows that 18F2 is specific for MP98 but that other MPs are available to bind to Con A. Scale bar is 5 µm.
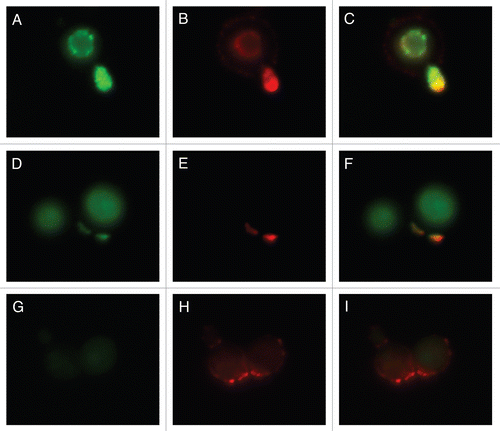
Table 1 Primers to make MP98(CDA2) deletion cassette and confirm gene replacement
Table 1 Primers to make CDA1 deletion cassette and confirm gene replacement