Figures & data
Figure 1. Effect of human serum/LBP on LPS/LTA-induced cytokine secretion. Cytokine secretion was measured after stimulation using either 100 ng/ml LPS or 10 μg/ml of LTA in the presence and absence of human serum or LBP. TNF-α was measured in the cell supernatant using a flow cytometric cytokine bead array system (Becton Dickinson). The results are representative from three independent experiments. Asterisks denote statistical significance (p < 0.001).
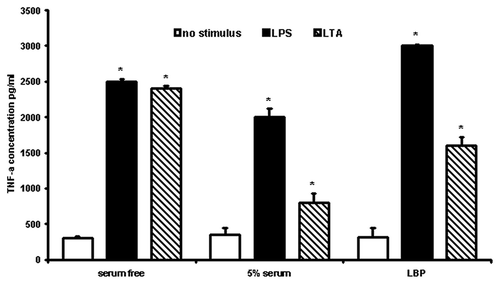
Figure 2. Two-dimensional gel electrophoresis of LPS/LTA-binding serum proteins. LPS (A) or LTA (B) was covalently coupled to NHS-activated columns. Normal human serum was passed through the columns. Unbound proteins were washed away, bound proteins were eluted, concentrated and analyzed by two-dimensional gel electrophoresis. The proteins that were excised for tryptic digestion are indicated by the numbers.
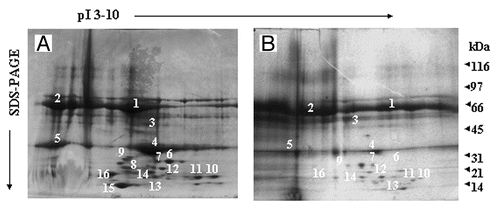
Figure 3. Mass spec analysis of excised proteins. Proteins of interest were excised, tryptic digested and analyzed by MALDI. Peptide profiles of albumin (A), LDL (B), Hemoglobin (C) and Apolipoprotein (D) are shown.
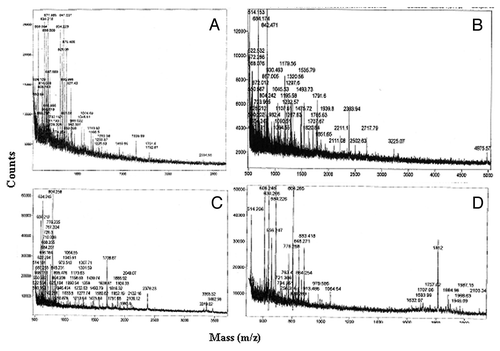
Figure 4. LPS/LTA-binding on human monocytes in the presence and absence of serum proteins. Human monocytes were incubated with either 100ng/ml FITC-LPS or 10 μg/ml of FITC-LTA in the presence and absence of different serum proteins (1 mg/ml). Fluorescence was detected by flow cytometry using a FACSCalibur (Becton Dickinson), counting 10,000 cells, not gated. T-test was performed comparing results against serum free values for LPS+ and LTA+, respectively. * and # denote statistical significance (p < 0.05). The results are representative from three independent experiments.
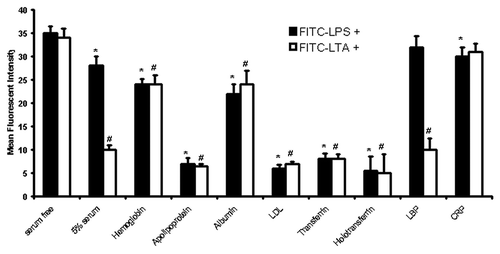
Figure 5. LPS/LTA-induced cytokine secretion in the presence and absence of different serum proteins. Human monocytes were incubated with 1 mg/ml of different serum proteins prior to incubation with either 100 ng/ml LPS or 10 μg/ml of LTA. Supernatant was collected and analyzed for TNF-α using a flow cytometric (Th1/Th2) bead array system (Becton Dickinson). T-test was performed comparing results against serum free values for LPS+ and LTA+, respectively. * and # denote statistical significance (p < 0.05). The results are representative from three independent experiments.
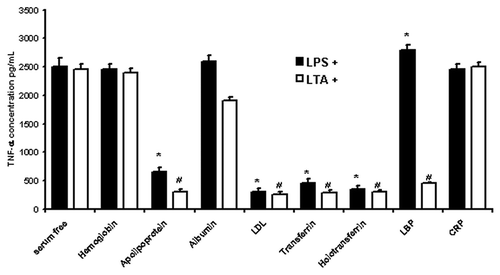
Table 1. Septic patients: microbiological findings, APACHE and SOFA scores and outcome (Mean ± SD)
Figure 6. Two-dimensional gel electrophoresis of LPS/LTA-binding serum proteins from septic serum. LPS or LTA was covalently coupled to NHS-activated columns. Human serum obtained from septic patients was passed through the columns. Unbound proteins were washed away, bound proteins were eluted, concentrated and analyzed by two-dimensional gel electrophoresis. Proteins of interest were excised for tryptic digestion and mass spec analysis. Twenty five samples were analyzed; this is a representative gel.
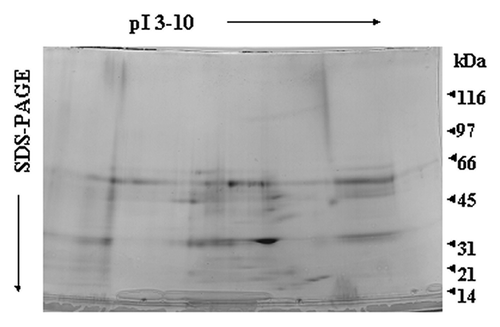
Figure 7. Differences in serum proteins from septic patients and normal controls. Relative integrated pixel intensity of each spot was quantitated by densitometry (Bio-Rad). The white bars represent mean protein quantitation from normal human serum, and the black bars are from septic serum. Asterisks denote significance (p < 0.05).
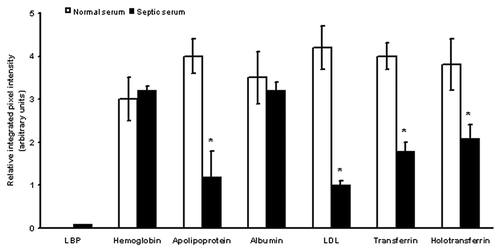
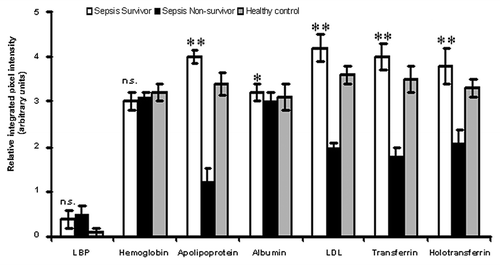
Figure 8. Comparison of proteomic profiles of sepsis survivors, non-survivors and healthy volunteers. Relative integrated pixel intensity of individual serum proteins from the proteomic profiles of sepsis survivors (white bar-charts), non-survivors (black bar-charts) or healthy volunteers (gray bar-charts) was quantitated by densitometry (Bio-Rad). **p < 0.001; *p < 0.05; n.s., not significant.