Figures & data
Figure 1 The role of YjeK and PoxA in the modification of EF-P. (A) A model for the post-translational modification of the conserved lysyl 34 residue of EF-P by the PoxA and YjeK enzymes. As shown the YjeK enzyme converts L-lysine to (R)-β-lysine, which is subsequently ligated onto EF-P to generate the lysyl-β-lysine modification, although the order of these two enzymes in this pathway could be reversed. The modified elongation factor subsequently interacts with the ribosome and the initiator tRNA to facilitate the formation of the first peptide bond of specific transcripts. The mechanism underlying why only certain transcripts are regulated by EF-P remains unclear. (B) The three dimensional structures of tRNA (left, PDB 1EHZ) and EF-P (right, PDB 1UEB)Citation24 reveal their similar overall structure. The sites of their modification by LysRS or by the aaRS paralog, PoxA, are indicated with arrows.
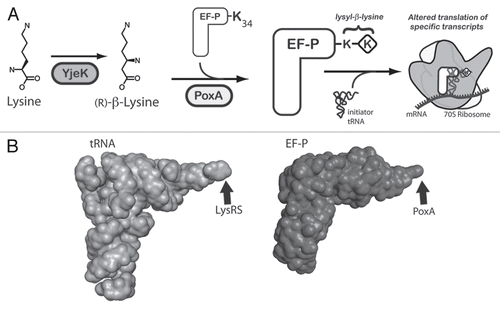
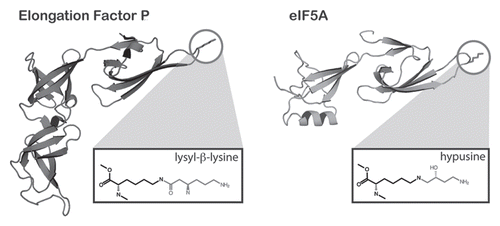
Figure 2 The structural similarity of EF-P and its modification to eIF5A. The structure of EF-P (left) is simliar to that of eIF5A of S. cerevisiae (PDB 3ER0) except that EF-P contains an additional C-terminal domain that is absent from eIF5A. Both factors contain a highly conserved loop, containing the modified lysine (circled). The structures of the modified amino acids are shown in boxes below each structure. The modification on EF-P is proposed to be lysyl-β-lysine although its structure has not been formally determined in the context of the native protein.