Figures & data
Figure 1 Replication of S. Typhimurium SPI2 mutants in epithelial cells. (A) HeLa cells were infected with exponentially-growing Salmonella, with a multiplicity of infection of 14. (B) CaCO2 cells were infected with exponentially-growing Salmonella, with a multiplicity of infection of 10. Error bars represent standard errors of the means. Fold replication was determined by comparing bacterial counts at 2 and 6 h post-infection.
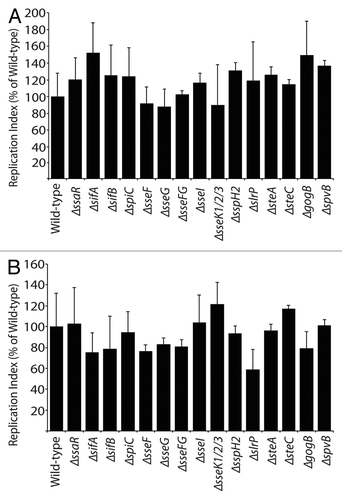
Figure 2 Replication of S. Typhimurium SPI2 mutants in RAW264.7 macrophages. Macrophages were infected with Salmonella, with a multiplicity of infection of 10. Fold replication was determined by comparing bacterial counts at 3 and 24 h post-infection. Error bars represent standard errors of the means. Asterisks indicate p < 0.05.
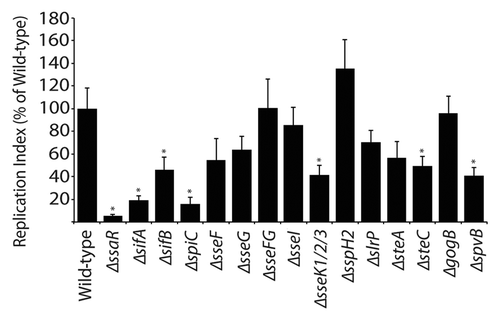
Figure 3 Bacterial counts of S. Typhimurium SPI2 mutants recovered from intestinal and systemic organs of C57BL/6 mice after oral infection. Mice were infected via oral gavage and sacrificed 5 d post-infection. Organs collected include ileum, cecum, colon, liver, spleen and mesenteric lymph nodes (MLN). Counts are given as colony-forming units (CFU) per milligram of tissue. Error bars represent standard deviation from the means. A minimum of 5 mice was used for each SPI2 mutant. Asterisks indicate **p < 0.001 or *p < 0.05.
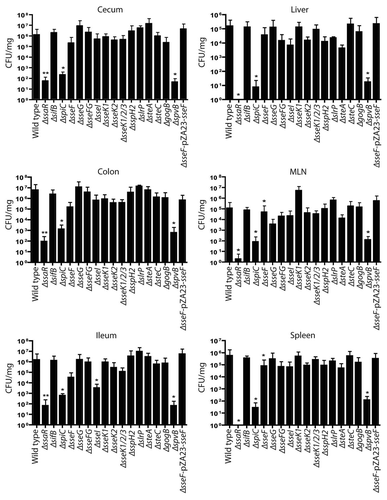
Figure 4 Bacterial counts of S. Typhimurium SPI2 mutants recovered from bile of C57BL/6 mice after oral infection. Mice were infected via oral gavage, sacrificed 5 d post-infection, and bile was extracted from gallbladders. Counts are given as colony-forming units (CFU) per microliter of bile. Error bars represent standard errors of the means. A minimum of 5 mice was used for each SPI2 mutant.
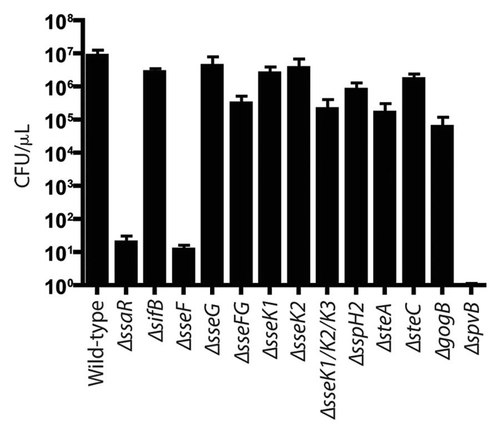
Figure 5 Bacterial count of S. Typhimurium SPI2 mutants recovered from systemic and intestinal organs of C57BL/6 mice 3 d post-infection. Mice were infected via oral gavage. Counts given represent colony forming units per milligram of tissue. Error bars represent standard errors from the means, each group contained 5 mice. Asterisks indicates *p < 0.05. Mesenteric lymph nodes (MLN).
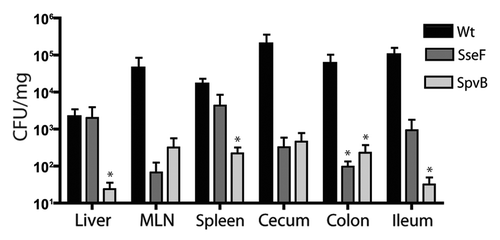
Table 1 Summary table of SPI2 mutants tested in RAW264.7 macrophage cells, HeLa and CaCo2 epithelial cells, and the murine typhoid fever model
Table 2 Bacterial strains and plasmids used in this study
Table 3 Oligonucleotides used to construct SPI2 gene deletions